8D4P
 
 | |
1SDI
 
 | | 1.65 A structure of Escherichia coli ycfC gene product | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETIC ACID, Hypothetical protein ycfC | | Authors: | Borek, D, Otwinowski, Z, Chen, Y, Skarina, T, Savchenko, A, Edwards, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-13 | | Release date: | 2004-08-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural analysis of Escherichia coli ycfC gene product
To be Published
|
|
6UN3
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with ticarcillin | | Descriptor: | (2R,4S)-2-[(1R)-1-{[(2R)-2-carboxy-2-(thiophen-3-yl)acetyl]amino}-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CALCIUM ION, GLYCEROL, ... | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2019-10-10 | | Release date: | 2019-10-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Influence of the alpha-Methoxy Group on the Reaction of Temocillin with Pseudomonas aeruginosa PBP3 and CTX-M-14 beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2019
|
|
6UN1
 
 | | Crystal structure of Pseudomonas aeruginosa PBP3 in complex with temocillin | | Descriptor: | (2R,4S)-2-[(1S)-1-{[(2R)-2-carboxy-2-(thiophen-3-yl)acetyl]amino}-1-methoxy-2-oxoethyl]-5,5-dimethyl-1,3-thiazolidine-4 -carboxylic acid, Peptidoglycan D,D-transpeptidase FtsI | | Authors: | Sacco, M, Chen, Y. | | Deposit date: | 2019-10-10 | | Release date: | 2019-10-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Influence of the alpha-Methoxy Group on the Reaction of Temocillin with Pseudomonas aeruginosa PBP3 and CTX-M-14 beta-Lactamase.
Antimicrob.Agents Chemother., 64, 2019
|
|
5LSW
 
 | | A CAF40-binding motif facilitates recruitment of the CCR4-NOT complex to mRNAs targeted by Drosophila Roquin | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell differentiation protein RCD1 homolog, LD12033p, ... | | Authors: | Sgromo, A, Raisch, T, Bawankar, P, Bhandari, D, Chen, Y, Kuzuoglu-Ozturk, D, Weichenrieder, O, Izaurralde, E. | | Deposit date: | 2016-09-05 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A CAF40-binding motif facilitates recruitment of the CCR4-NOT complex to mRNAs targeted by Drosophila Roquin.
Nat Commun, 8, 2017
|
|
2QGU
 
 | | Three-dimensional structure of the phospholipid-binding protein from Ralstonia solanacearum Q8XV73_RALSQ in complex with a phospholipid at the resolution 1.53 A. Northeast Structural Genomics Consortium target RsR89 | | Descriptor: | DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Probable signal peptide protein | | Authors: | Kuzin, A.P, Chen, Y, Jayaraman, S, Chen, C.X, Fang, Y, Cunningham, K, Ma, L.-C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-07-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-dimensional structure of the phospholipid-binding protein from Ralstonia solanacearum Q8XV73_RALSQ in complex with a phospholipid at the resolution 1.53 A.
To be Published
|
|
6LHD
 
 | | Crystal structure of p53/BCL-xL fusion complex | | Descriptor: | ZINC ION, fusion protein of Bcl-2-like protein 1 and Isoform 6 of Cellular tumor antigen p53 | | Authors: | Wei, H, Chen, Y. | | Deposit date: | 2019-12-07 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structural insight into the molecular mechanism of p53-mediated mitochondrial apoptosis.
Nat Commun, 12, 2021
|
|
1WPB
 
 | | Structure of Escherichia coli yfbU gene product | | Descriptor: | CHLORIDE ION, GLYCEROL, hypothetical protein yfbU | | Authors: | Borek, D, Chen, Y, Zheng, M, Skarina, T, Savchenko, A, Edwards, A, Otwinowski, Z, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-09-01 | | Release date: | 2004-12-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Escherichia coli yfbU gene product
To be Published
|
|
4Z8I
 
 | | Crystal structure of Branchiostoma belcheri tsingtauense peptidoglycan recognition protein 3 | | Descriptor: | ZINC ION, peptidoglycan recognition protein 3 | | Authors: | Wang, W.J, Cheng, W, Jiang, Y.L, Luo, M, Cao, D.D, Chi, C.B, Yang, H.B, Chen, Y, Zhou, C.Z. | | Deposit date: | 2015-04-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Activity Augmentation of Amphioxus Peptidoglycan Recognition Protein BbtPGRP3 via Fusion with a Chitin Binding Domain
Plos One, 10, 2015
|
|
2D2R
 
 | | Crystal structure of Helicobacter pylori Undecaprenyl Pyrophosphate Synthase | | Descriptor: | Undecaprenyl Pyrophosphate Synthase | | Authors: | Kuo, C.J, Guo, R.T, Chen, C.L, Ko, T.P, Cheng, Y.S, Cheng, Y.L, Liang, P.H, Wang, A.H.-J. | | Deposit date: | 2005-09-16 | | Release date: | 2006-09-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure-based inhibitors exhibit differential activities against Helicobacter pylori and Escherichia coli undecaprenyl pyrophosphate synthases.
J.Biomed.Biotechnol., 2008, 2008
|
|
5WSU
 
 | | Crystal structure of Myosin VIIa IQ5-SAH in complex with apo-CaM | | Descriptor: | Calmodulin, Unconventional myosin-VIIa | | Authors: | Li, J, Chen, Y, Deng, Y, Lu, Q, Zhang, M. | | Deposit date: | 2016-12-08 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Ca(2+)-Induced Rigidity Change of the Myosin VIIa IQ Motif-Single alpha Helix Lever Arm Extension
Structure, 25, 2017
|
|
5WSV
 
 | | Crystal structure of Myosin VIIa IQ5 in complex with Ca2+-CaM | | Descriptor: | CALCIUM ION, Calmodulin, SULFATE ION, ... | | Authors: | Li, J, Chen, Y, Deng, Y, Lu, Q, Zhang, M. | | Deposit date: | 2016-12-08 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Ca(2+)-Induced Rigidity Change of the Myosin VIIa IQ Motif-Single alpha Helix Lever Arm Extension
Structure, 25, 2017
|
|
6BGJ
 
 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in LMNG | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, L, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
2QIK
 
 | | Crystal structure of YkqA from Bacillus subtilis. Northeast Structural Genomics Target SR631 | | Descriptor: | CITRIC ACID, UPF0131 protein ykqA | | Authors: | Benach, J, Chen, Y, Forouhar, F, Seetharaman, J, Baran, M.C, Cunningham, K, Ma, L.-C, Owens, L, Chen, C.X, Rong, X, Janjua, H, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-07-05 | | Release date: | 2007-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of YkqA from Bacillus subtilis.
To be Published
|
|
5WST
 
 | | Crystal structure of Myo7a SAH | | Descriptor: | Unconventional myosin-VIIa | | Authors: | Li, J, Chen, Y, Deng, Y, Lu, Q, Zhang, M. | | Deposit date: | 2016-12-08 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ca(2+)-Induced Rigidity Change of the Myosin VIIa IQ Motif-Single alpha Helix Lever Arm Extension
Structure, 25, 2017
|
|
6BGI
 
 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in nanodisc | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, J, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
7BWQ
 
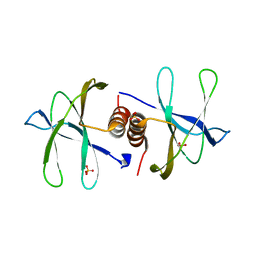 | | Structure of nonstructural protein Nsp9 from SARS-CoV-2 | | Descriptor: | Nsp9, SULFATE ION | | Authors: | Zhang, C, Chen, Y, Li, L, Su, D. | | Deposit date: | 2020-04-15 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural basis for the multimerization of nonstructural protein nsp9 from SARS-CoV-2.
Mol Biomed, 1, 2020
|
|
5GZZ
 
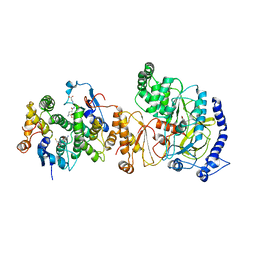 | | Crystal Structure of FIN219-SjGST complex with JA | | Descriptor: | GLUTATHIONE, Glutathione S-transferase class-mu 26 kDa isozyme, Jasmonic acid-amido synthetase JAR1, ... | | Authors: | Chen, C.Y, Cheng, Y.S. | | Deposit date: | 2016-10-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.386 Å) | | Cite: | Structural basis of jasmonate-amido synthetase FIN219 in complex with glutathione S-transferase FIP1 during the JA signal regulation
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
4X3Q
 
 | | Crystal structure of S-adenosylmethionine-dependent methyltransferase SibL in complex with SAH | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SibL | | Authors: | liu, J.S, Chen, S.C, Yang, C.S, Huang, C.H, Chen, Y. | | Deposit date: | 2014-12-01 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.586 Å) | | Cite: | Crystal structure of S-adenosylmethionine-dependent methyltransferase SibL in complex with SAH
To Be Published, 2015
|
|
2MLK
 
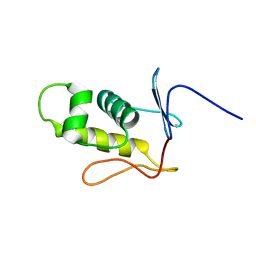 | | Three-dimensional structure of the C-terminal DNA-binding domain of RstA protein from Klebsiella pneumoniae | | Descriptor: | RstA | | Authors: | Fang, P, Chen, S, Cheng, Y, Chang, C, Yu, T, Huang, T. | | Deposit date: | 2014-03-02 | | Release date: | 2014-07-16 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural dynamics of the two-component response regulator RstA in recognition of promoter DNA element.
Nucleic Acids Res., 42, 2014
|
|
1JJR
 
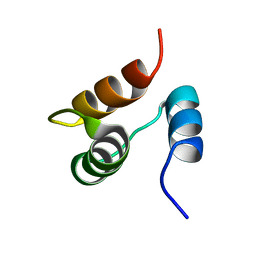 | |
6L6Q
 
 | |
7MN2
 
 | | Rules for designing protein fold switches and their implications for the folding code | | Descriptor: | Sb2 | | Authors: | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
7MP7
 
 | | Rules for designing protein fold switches and their implications for the folding code | | Descriptor: | Sb3 | | Authors: | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | Deposit date: | 2021-05-04 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
7MN1
 
 | | Rules for designing protein fold switches and their implications for the folding code | | Descriptor: | Sa1 | | Authors: | He, Y, Chen, Y, Ruan, B, Choi, J, Chen, Y, Motabar, D, Solomon, T, Simmerman, R, Kauffman, T, Gallagher, T, Bryan, P, Orban, J. | | Deposit date: | 2021-04-30 | | Release date: | 2022-05-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design and characterization of a protein fold switching network.
Nat Commun, 14, 2023
|
|
