3VPM
 
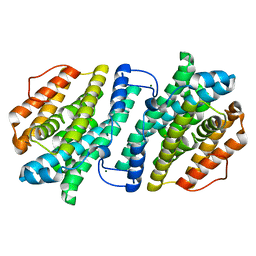 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human ribonucleotide reductase M2 subunit
To be Published
|
|
3VPO
 
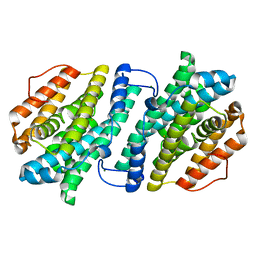 | | Crystal structure of human ribonucleotide reductase subunit M2 (hRRM2) mutant | | Descriptor: | FE (III) ION, MAGNESIUM ION, Ribonucleoside-diphosphate reductase subunit M2 | | Authors: | Chen, X, Xu, Z, Liu, H, Zhang, L, Chen, B, Zhu, L, Yang, C, Zhu, W, Shao, J. | | Deposit date: | 2012-03-05 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Essential role of E106 in the proton-coupled electron transfer in human
to be published
|
|
8QH2
 
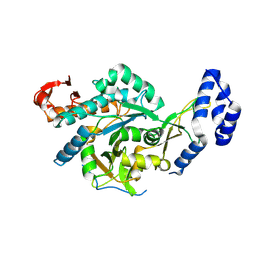 | | Crystal structure of chimeric UAP1L1 | | Descriptor: | UDP-N-acetylhexosamine pyrophosphorylase,UDP-N-acetylhexosamine pyrophosphorylase-like protein 1 | | Authors: | Chen, X, Yan, K, Bartual, S, van Aalten, D. | | Deposit date: | 2023-09-06 | | Release date: | 2024-09-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | UAP1L1 is an active pyrophosphorylase with an active site disulfide regulating protein thermostability
To Be Published
|
|
1QAW
 
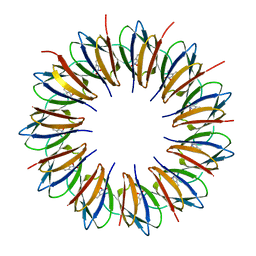 | | Regulatory Features of the TRP Operon and the Crystal Structure of the TRP RNA-Binding Attenuation Protein from Bacillus Stearothermophilus. | | Descriptor: | TRP RNA-BINDING ATTENUATION PROTEIN, TRYPTOPHAN | | Authors: | Chen, X.-P, Antson, A.A, Yang, M, Baumann, C, Dodson, E.J, Dodson, G.G, Gollnick, P. | | Deposit date: | 1999-03-31 | | Release date: | 1999-04-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Regulatory features of the trp operon and the crystal structure of the trp RNA-binding attenuation protein from Bacillus stearothermophilus.
J.Mol.Biol., 289, 1999
|
|
1EKD
 
 | | NMR AND MOLECULAR MODELING REVEAL THAT DIFFERENT HYDROGEN BONDING PATTERNS ARE POSSIBLE FOR GU PAIRS: ONE HYDROGEN BOND FOR EACH GU PAIR IN R(GGCGUGCC)2 AND TWO FOR EACH GU PAIR IN R(GAGUGCUC)2 | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*UP*GP*CP*C)-3') | | Authors: | Chen, X, McDowell, J.A, Kierzek, R, Krugh, T.R, Turner, D.H. | | Deposit date: | 2000-03-07 | | Release date: | 2000-11-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G.U pairs: one hydrogen bond for each G.U pair in r(GGCGUGCC)(2) and two for each G.U pair in r(GAGUGCUC)(2).
Biochemistry, 39, 2000
|
|
6OEP
 
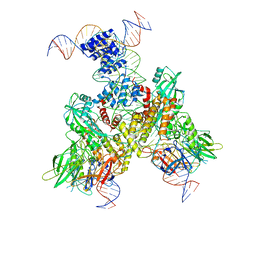 | | Cryo-EM structure of mouse RAG1/2 12RSS-NFC/23RSS-PRC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OET
 
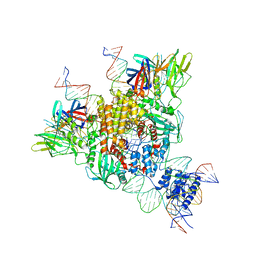 | | Cryo-EM structure of mouse RAG1/2 STC complex | | Descriptor: | CALCIUM ION, DNA (30-MER), DNA (39-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-22 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | How mouse RAG recombinase avoids DNA transposition.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6OEN
 
 | | Cryo-EM structure of mouse RAG1/2 PRC complex (DNA1) | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (57-MER), ... | | Authors: | Chen, X, Cui, Y, Zhou, Z.H, Yang, W, Gellert, M. | | Deposit date: | 2019-03-27 | | Release date: | 2020-01-29 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cutting antiparallel DNA strands in a single active site.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2OZ4
 
 | | Structural Plasticity in IgSF Domain 4 of ICAM-1 Mediates Cell Surface Dimerization | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB FRAGMENT LIGHT CHAIN, ... | | Authors: | Chen, X, Kim, T.D, Carman, C.V, Mi, L, Song, G, Springer, T.A. | | Deposit date: | 2007-02-23 | | Release date: | 2007-10-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural plasticity in Ig superfamily domain 4 of ICAM-1 mediates cell surface dimerization.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
8YZ5
 
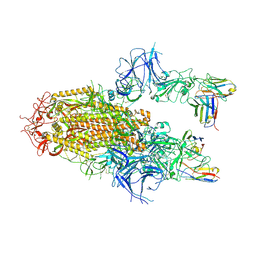 | | SARS-CoV-2 Delta Spike in complex with Fab of JE-5C | | Descriptor: | Fab heavy chain of JE-5C, Fab light chain of JE-5C, Spike glycoprotein | | Authors: | Chen, X, Wu, Y.-M. | | Deposit date: | 2024-04-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
8YZ6
 
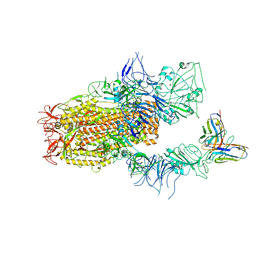 | | SARS-CoV-2 Spike (BA.1) in complex with Fab of JH-8B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab heavy chain of JH-8B, ... | | Authors: | Chen, X, Wu, Y.-M. | | Deposit date: | 2024-04-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.55 Å) | | Cite: | The presence of broadly neutralizing anti-SARS-CoV-2 RBD antibodies elicited by primary series and booster dose of COVID-19 vaccine.
Plos Pathog., 20, 2024
|
|
5DIP
 
 | | Crystal structure of lpg0406 in reduced form from Legionella pneumophila | | Descriptor: | Alkyl hydroperoxide reductase AhpD, GLYCEROL, SODIUM ION | | Authors: | Chen, X, Gong, X, Zhang, N, Ge, H. | | Deposit date: | 2015-09-01 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structure of lpg0406, a carboxymuconolactone decarboxylase family protein possibly involved in antioxidative response from Legionella pneumophila
Protein Sci., 24, 2015
|
|
8ZF1
 
 | | Crystal structure of a Chemo Triplet Photoenzyme (CTPe) | | Descriptor: | N-(9-oxidanylidenethioxanthen-2-yl)ethanamide, Transcriptional regulator, PadR-like family | | Authors: | Chen, X, Qian, J.Y, Guo, J, Wu, Y.Z, Zhong, F.R. | | Deposit date: | 2024-05-07 | | Release date: | 2024-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Chemogenetic Evolution of Diversified Photoenzymes for Enantioselective [2 + 2] Cycloadditions in Whole Cells.
J.Am.Chem.Soc., 146, 2024
|
|
7X9E
 
 | | Crystal structure of the 76E1 Fab in complex with a SARS-CoV-2 spike peptide | | Descriptor: | 76E1 Fab Heavy Chain, 76E1 Fab Light Chain, Spike peptide | | Authors: | Chen, X, Zhang, T, Ding, J, Sun, X, Sun, B. | | Deposit date: | 2022-03-15 | | Release date: | 2022-05-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Neutralization mechanism of a human antibody with pan-coronavirus reactivity including SARS-CoV-2.
Nat Microbiol, 7, 2022
|
|
7AUC
 
 | |
6XBU
 
 | | polymerase domain of polymerase-theta | | Descriptor: | 2'-3'-DIDEOXYGUANOSINE-5'-TRIPHOSPHATE, DNA (5'-D(P*GP*TP*CP*AP*TP*TP*G)-3'), DNA polymerase theta, ... | | Authors: | Chen, X, Pomerantz, R, Zhao, J. | | Deposit date: | 2020-06-07 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Pol theta reverse transcribes RNA and promotes RNA-templated DNA repair.
Sci Adv, 7, 2021
|
|
5YQG
 
 | | The structure of 14-3-3 and pNumb peptide | | Descriptor: | 14-3-3 protein eta, Peptide from Protein numb homolog | | Authors: | Chen, X, Liu, Z, Wen, W. | | Deposit date: | 2017-11-06 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural determinants controlling 14-3-3 recruitment to the endocytic adaptor Numb and dissociation of the Numb/alpha-adaptin complex.
J. Biol. Chem., 293, 2018
|
|
7AUD
 
 | |
1G3A
 
 | |
8KFO
 
 | | Crystal structure of BSA in complex with B3 | | Descriptor: | 6-[(~{E})-2-[1-[2-[2-(2-methoxyethoxy)ethoxy]ethyl]pyridin-1-ium-4-yl]ethenyl]-~{N},~{N}-dimethyl-naphthalen-2-amine, Albumin | | Authors: | Chen, X, Ge, Y.H, Yang, H, Fang, B, Li, L. | | Deposit date: | 2023-08-16 | | Release date: | 2024-08-21 | | Last modified: | 2025-03-05 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Albumins constrainting the conformation of mitochondria-targeted photosensitizers for tumor-specific photodynamic therapy.
Biomaterials, 315, 2025
|
|
2Z59
 
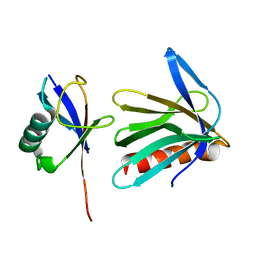 | | Complex Structures of Mouse Rpn13 (22-130aa) and ubiquitin | | Descriptor: | Protein ADRM1, Ubiquitin | | Authors: | Chen, X, Schreiner, P, Groll, M, Walters, K.J. | | Deposit date: | 2007-07-01 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction.
Nature, 453, 2008
|
|
5ZXF
 
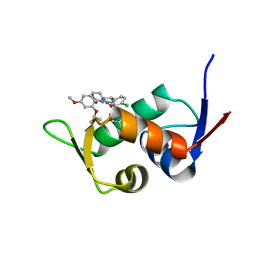 | | The 1.25A Crystal structure of His6-tagged Mdm2 in complex with nutlin-3a | | Descriptor: | 4-({(4S,5R)-4,5-bis(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Cheng, X.Y, Su, Z.D, Pi, N, Cao, C.Z, Zhao, Z.T, Zhou, J.J, Chen, R, Kuang, Z.K, Huang, Y.Q. | | Deposit date: | 2018-05-19 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The 1.25A Crystal structure of His6-tagged Mdm2 in complex with nutlin-3a
To Be Published
|
|
6Z2F
 
 | |
6IG5
 
 | |
6RWG
 
 | | Structure of HIV-1 CAcSP1NC mutant(W41A,M42A) interacting with maturation inhibitor EP39 | | Descriptor: | Gag polyprotein, ZINC ION | | Authors: | Chen, X, Coric, P, Larue, V, Nonin-Lecomte, S, Bouaziz, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-05 | | Release date: | 2020-05-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The HIV-1 maturation inhibitor, EP39, interferes with the dynamic helix-coil equilibrium of the CA-SP1 junction of Gag.
Eur.J.Med.Chem., 204, 2020
|
|
