5A7X
 
 | | negative stain EM of BG505 SOSIP.664 in complex with sCD4, 17b, and 8ANC195 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB OF BROADLY NEUTRALIZING ANTIBODY 17B, FAB OF BROADLY NEUTRALIZING ANTIBODY 8ANC195, ... | | Authors: | Scharf, L, Wang, H, Gao, H, Chen, S, McDowall, A, Bjorkman, P. | | Deposit date: | 2015-07-10 | | Release date: | 2015-08-05 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Broadly Neutralizing Antibody 8Anc195 Recognizes Closed and Open States of HIV-1 Env.
Cell(Cambridge,Mass.), 162, 2015
|
|
3JBL
 
 | | Cryo-EM Structure of the Activated NAIP2/NLRC4 Inflammasome Reveals Nucleated Polymerization | | Descriptor: | NLR family CARD domain-containing protein 4 | | Authors: | Zhang, L, Chen, S, Ruan, J, Wu, J, Tong, A.B, Yin, Q, Li, Y, David, L, Lu, A, Wang, W.L, Marks, C, Ouyang, Q, Zhang, X, Mao, Y, Wu, H. | | Deposit date: | 2015-09-05 | | Release date: | 2015-10-21 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization.
Science, 350, 2015
|
|
5BR4
 
 | | E. coli lactaldehyde reductase (FucO) M185C mutant | | Descriptor: | CHLORIDE ION, GLYCEROL, Lactaldehyde reductase, ... | | Authors: | Cahn, J.K.B, Brinkmann-Chen, S, Arnold, F.H. | | Deposit date: | 2015-05-29 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (0.91 Å) | | Cite: | Mutations in adenine-binding pockets enhance catalytic properties of NAD(P)H-dependent enzymes.
Protein Eng.Des.Sel., 29, 2016
|
|
2H53
 
 | | Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein Hsp26 | | Descriptor: | small heat shock protein Hsp26 | | Authors: | White, H.E, Orlova, E.V, Chen, S, Wang, L, Ignatiou, A, Gowen, B, Stromer, T, Franzmann, T.M, Haslbeck, M, Buchner, J, Saibil, H.R. | | Deposit date: | 2006-05-25 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein hsp26
Structure, 14, 2006
|
|
2H50
 
 | | Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein Hsp26 | | Descriptor: | small heat shock protein Hsp26 | | Authors: | White, H.E, Orlova, E.V, Chen, S, Wang, L, Ignatiou, A, Gowen, B, Stromer, T, Franzmann, T.M, Haslbeck, M, Buchner, J, Saibil, H.R. | | Deposit date: | 2006-05-25 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (10.8 Å) | | Cite: | Multiple distinct assemblies reveal conformational flexibility in the small heat shock protein hsp26
Structure, 14, 2006
|
|
7DB7
 
 | | Crystal structure of Mycobacterium tuberculosis phenylalanyl-tRNA synthetase in complex with compound GDI05-001 | | Descriptor: | 1-[3-[2-(1H-indol-3-yl)ethylsulfamoyl]phenyl]-3-(1,3-thiazol-2-yl)urea, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit, ... | | Authors: | Xu, M, Zhang, X, Xu, L, Chen, S. | | Deposit date: | 2020-10-19 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Re-discovery of PF-3845 as a new chemical scaffold inhibiting phenylalanyl-tRNA synthetase in Mycobacterium tuberculosis .
J.Biol.Chem., 2021
|
|
7DAW
 
 | | Crystal structure of Mycobacterium tuberculosis phenylalanyl-tRNA synthetase | | Descriptor: | Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit, SULFATE ION | | Authors: | Xu, M, Zhang, X, Xu, L, Chen, S. | | Deposit date: | 2020-10-18 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Re-discovery of PF-3845 as a new chemical scaffold inhibiting phenylalanyl-tRNA synthetase in Mycobacterium tuberculosis .
J.Biol.Chem., 2021
|
|
7DB8
 
 | | Crystal structure of Mycobacterium tuberculosis phenylalanyl-tRNA synthetase in complex with compound PF-3845 | | Descriptor: | N-pyridin-3-yl-4-[[3-[5-(trifluoromethyl)pyridin-2-yl]oxyphenyl]methyl]piperidine-1-carboxamide, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit, ... | | Authors: | Xu, M, Zhang, X, Xu, L, Chen, S. | | Deposit date: | 2020-10-19 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Re-discovery of PF-3845 as a new chemical scaffold inhibiting phenylalanyl-tRNA synthetase in Mycobacterium tuberculosis .
J.Biol.Chem., 2021
|
|
3QB5
 
 | | Human C3PO complex in the presence of MnSO4 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Ye, X, Huang, N, Liu, Y, Paroo, Z, Chen, S, Zhang, H, Liu, Q. | | Deposit date: | 2011-01-12 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of C3PO and mechanism of human RISC activation.
Nat.Struct.Mol.Biol., 18, 2011
|
|
2MOA
 
 | |
3TCM
 
 | | Crystal Structure of Alanine Aminotransferase from Hordeum vulgare | | Descriptor: | Alanine aminotransferase 2, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE | | Authors: | Rydel, T.J, Sturman, E.J, Halls, C, Chen, S, Zeng, J, Evdokimov, A, Duff, S.M.G. | | Deposit date: | 2011-08-09 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The Enzymology of alanine aminotransferase (AlaAT) isoforms from Hordeum vulgare and other organisms, and the HvAlaAT crystal structure.
Arch.Biochem.Biophys., 528, 2012
|
|
7X4E
 
 | | Structure of 10635-DndE | | Descriptor: | DNA sulfur modification protein DndE, GLYCEROL | | Authors: | Haiyan, G, Wei, H, Chen, S, Wang, L, Wu, G. | | Deposit date: | 2022-03-02 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural and Functional Analysis of DndE Involved in DNA Phosphorothioation in the Haloalkaliphilic Archaea Natronorubrum bangense JCM10635.
Mbio, 13, 2022
|
|
8IAH
 
 | | Structure of mammalian spectrin-actin junctional complex of membrane skeleton, State I, Global map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Li, N, Chen, S, Gao, N. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of membrane skeleton organization in red blood cells.
Cell, 186, 2023
|
|
8IAI
 
 | | Structure of mammalian spectrin-actin junctional complex of membrane skeleton, State II, Global map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Li, N, Chen, S, Gao, N. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of membrane skeleton organization in red blood cells.
Cell, 186, 2023
|
|
8IB2
 
 | | Structure of mammalian spectrin-actin junctional complex of membrane skeleton, Pointed-end segment, headpiece domain of dematin optimized | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Li, N, Chen, S, Gao, N. | | Deposit date: | 2023-02-09 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of membrane skeleton organization in red blood cells.
Cell, 186, 2023
|
|
4GLY
 
 | | Human urokinase-type plasminogen activator uPA in complex with the two-disulfide bridge peptide UK504 | | Descriptor: | BICYCLIC PEPTIDE INHIBITOR UK504, CHLORIDE ION, GLYCEROL, ... | | Authors: | Buth, S.A, Leiman, P.G, Chen, S, Heinis, C. | | Deposit date: | 2012-08-15 | | Release date: | 2013-05-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.518 Å) | | Cite: | Bicyclic Peptide Ligands Pulled out of Cysteine-Rich Peptide Libraries.
J.Am.Chem.Soc., 135, 2013
|
|
4GPO
 
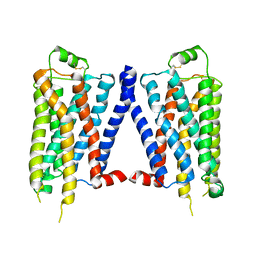 | |
5E4R
 
 | | Crystal structure of domain-duplicated synthetic class II ketol-acid reductoisomerase 2Ia_KARI-DD | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Ketol-acid reductoisomerase, ... | | Authors: | Cahn, J.K.B, Brinkmann-Chen, S, Buller, A.R, Arnold, F.H. | | Deposit date: | 2015-10-07 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Artificial domain duplication replicates evolutionary history of ketol-acid reductoisomerases.
Protein Sci., 25, 2016
|
|
5FNA
 
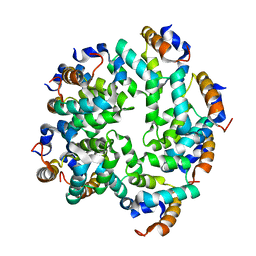 | | Cryo-EM reconstruction of caspase-1 CARD | | Descriptor: | Caspase-1 | | Authors: | Li, Y, Lu, A, Schmidt, F.I, Yin, Q, Chen, S, Fu, T.M, Tong, A.B, Ploegh, H.L, Mao, Y, Wu, H. | | Deposit date: | 2015-11-11 | | Release date: | 2016-03-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Molecular Basis of Caspase-1 Polymerization and its Inhibition by a Novel Capping Mechanism
Nat.Struct.Mol.Biol., 23, 2016
|
|
4J1L
 
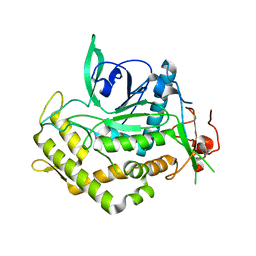 | | Mutant Endotoxin TeNT | | Descriptor: | Tetanus toxin, ZINC ION | | Authors: | Guo, J, Pan, X, Zhao, Y, Chen, S. | | Deposit date: | 2013-02-01 | | Release date: | 2014-02-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineering Clostridia Neurotoxins with elevated catalytic activity
Toxicon, 74, 2013
|
|
5GOV
 
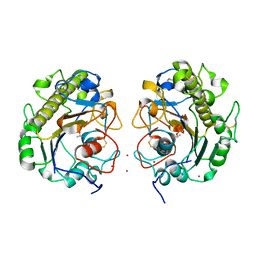 | | Crystal Structure of MCR-1, a phosphoethanolamine transferase, extracellular domain | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, ZINC ION | | Authors: | Hu, M, Guo, J, Chen, S, Hao, Q. | | Deposit date: | 2016-07-29 | | Release date: | 2016-12-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal Structure of Escherichia coli originated MCR-1, a phosphoethanolamine transferase for Colistin Resistance.
Sci Rep, 6, 2016
|
|
8PVI
 
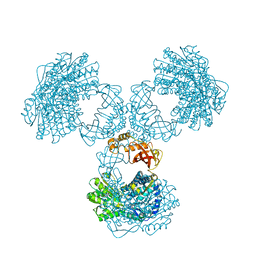 | | Structure of PaaZ determined by cryoEM at 100 keV | | Descriptor: | Bifunctional protein PaaZ | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PVG
 
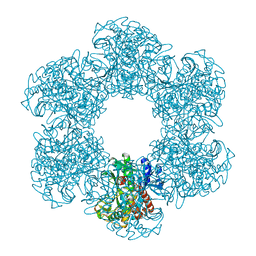 | | Structure of E. coli glutamine synthetase determined by cryoEM at 100 keV | | Descriptor: | Glutamine synthetase | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PVB
 
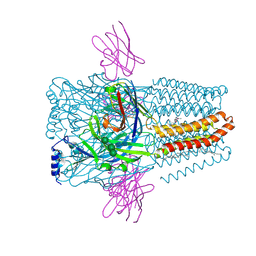 | | Structure of GABAAR determined by cryoEM at 100 keV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, DECANE, ... | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8PVA
 
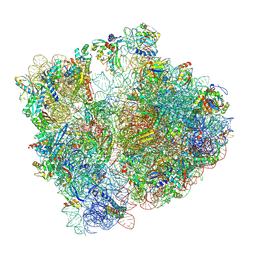 | | Structure of bacterial ribosome determined by cryoEM at 100 keV | | Descriptor: | 16S rRNA, 23S rRNA, 50S ribosomal protein L14, ... | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
