5L9C
 
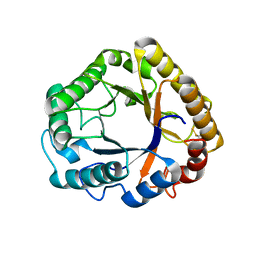 | | Crystal structure of an endoglucanase from Penicillium verruculosum in complex with cellobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Nemashkalov, V, Vakhrusheva, A, Tishchenko, S, Gabdulkhakov, A, Kravchenko, O, Gusakov, A, Sinisyn, A. | | Deposit date: | 2016-06-10 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of an endoglucanase from Penicillium verruculosum in complex with cellobiose
to be published
|
|
6TPC
 
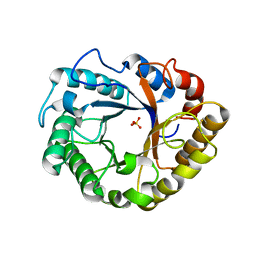 | | Crystal structure of Endoglucanase N194A from Penicillium verruculosum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endoglucanase, PHOSPHATE ION, ... | | Authors: | Nemashkalov, V, Kravchenko, O, Gabdulkhakov, A, Tischenko, S, Rozhkova, A, Sinitsyn, A. | | Deposit date: | 2019-12-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5221895 Å) | | Cite: | Crystal structure of Endoglucanase N194A from Penicillium verruculosum
To Be Published
|
|
8YLS
 
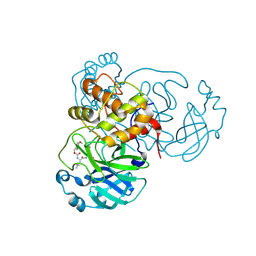 | | Structure of SARS-CoV-2 Mpro in complex with its degrader | | Descriptor: | (4-methoxyphenyl)methyl ~{N}-[(2~{S})-4-methyl-1-oxidanylidene-1-[[(2~{S})-1-oxidanylidene-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]pentan-2-yl]carbamate, 3C-like proteinase nsp5 | | Authors: | Feng, Y, Li, W, Cheng, S.H, Li, X.B. | | Deposit date: | 2024-03-06 | | Release date: | 2024-09-04 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | The Main Protease Degraders: Potent Broad-Spectrum Agents for Anti-Coronavirus Agents
To Be Published
|
|
5MKN
 
 | |
3QOY
 
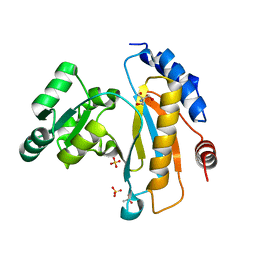 | | Crystal structure of ribosomal protein L1 from Aquifex aeolicus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 50S ribosomal protein L1, ACETIC ACID, ... | | Authors: | Gabdulkhakov, A.G, Tishchenko, S.V, Nikonova, E.U, Shkliaeva, A.A, Garber, M.B, Nikonov, S.V, Nevskaya, N.A. | | Deposit date: | 2011-02-11 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Ribosomal Protein L1 from the Bacterium Aquifex Aeolicus
Crystallography Reports, 56, 2011
|
|
8Q7Y
 
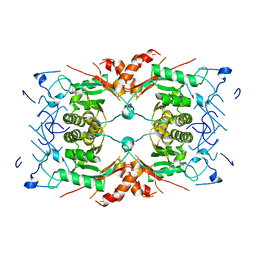 | |
6HD8
 
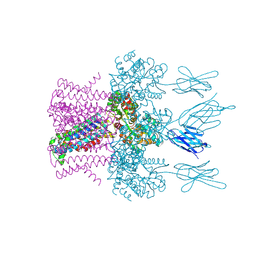 | | Crystal structure of the potassium channel MtTMEM175 in complex with a Nanobody-MBP fusion protein | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein, POTASSIUM ION, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
6HDB
 
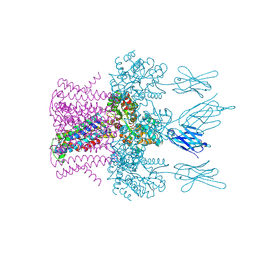 | | Crystal structure of the potassium channel MtTMEM175 with zinc | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein, POTASSIUM ION, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
6HD9
 
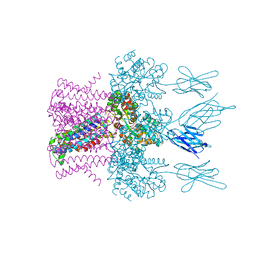 | | Crystal structure of the potassium channel MtTMEM175 with rubidium | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein, RUBIDIUM ION, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
6HDC
 
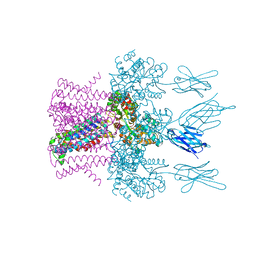 | | Crystal structure of the potassium channel MtTMEM175 T38A variant in complex with a Nanobody-MBP fusion protein | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein,Maltose/maltodextrin-binding periplasmic protein, POTASSIUM ION, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
5D3Q
 
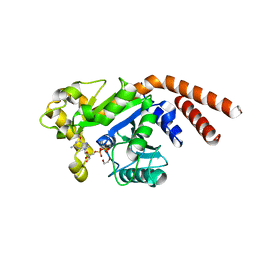 | | Dynamin 1 GTPase-BSE fusion dimer complexed with GDP | | Descriptor: | 1,2-ETHANEDIOL, Dynamin-1,Dynamin-1, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Anand, R, Eschenburg, S, Reubold, T.F. | | Deposit date: | 2015-08-06 | | Release date: | 2015-12-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the GTPase domain and the bundle signalling element of dynamin in the GDP state.
Biochem.Biophys.Res.Commun., 469, 2016
|
|
6HDA
 
 | | Crystal structure of the potassium channel MtTMEM175 with cesium | | Descriptor: | CESIUM ION, DODECYL-BETA-D-MALTOSIDE, Nanobody,Maltose/maltodextrin-binding periplasmic protein, ... | | Authors: | Brunner, J.D, Jakob, R.P, Schulze, T, Neldner, Y, Moroni, A, Thiel, G, Maier, T, Schenck, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for ion selectivity in TMEM175 K+channels.
Elife, 9, 2020
|
|
7TN9
 
 | | Structure of the Inmazeb cocktail and resistance to escape against Ebola virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein, GP2, ... | | Authors: | Rayaprolu, V, Fulton, B, Rafique, A, Arturo, E, Williams, D, Hariharan, C, Callaway, H, Parvate, A, Schendel, S.L, Parekh, D, Hui, S, Shaffer, K, Pascal, K.E, Wloga, E, Giordano, S, Copin, R, Franklin, M, Boytz, R.M, Donahue, C, Davey, R, Baum, A, Kyratsous, C.A, Saphire, E.O. | | Deposit date: | 2022-01-20 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the Inmazeb cocktail and resistance to Ebola virus escape.
Cell Host Microbe, 31, 2023
|
|
4WIT
 
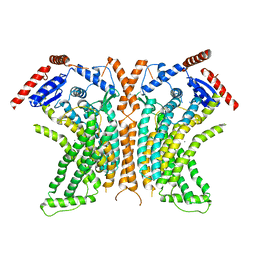 | | TMEM16 lipid scramblase in crystal form 2 | | Descriptor: | CALCIUM ION, Predicted protein | | Authors: | Dutzler, R, Brunner, J.D, Lim, N.K, Schenck, S. | | Deposit date: | 2014-09-26 | | Release date: | 2014-11-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | X-ray structure of a calcium-activated TMEM16 lipid scramblase.
Nature, 516, 2014
|
|
8QLN
 
 | |
4WIS
 
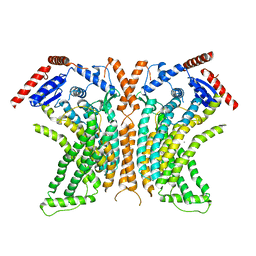 | | Crystal structure of the lipid scramblase nhTMEM16 in crystal form 1 | | Descriptor: | CALCIUM ION, lipid scramblase | | Authors: | Dutzler, R, Brunner, J.D, Lim, N.K, Schenck, S. | | Deposit date: | 2014-09-26 | | Release date: | 2014-11-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | X-ray structure of a calcium-activated TMEM16 lipid scramblase.
Nature, 516, 2014
|
|
4RWP
 
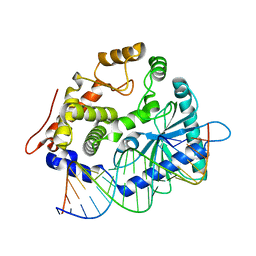 | | Crystal structure of porcine OAS1 in complex with dsRNA | | Descriptor: | 2'-5'-oligoadenylate synthase 1, RNA (5'-R(*GP*GP*CP*UP*UP*UP*UP*GP*AP*CP*CP*UP*UP*UP*AP*UP*GP*AP*A)-3'), RNA (5'-R(*UP*UP*CP*AP*UP*AP*AP*AP*GP*GP*UP*CP*AP*AP*AP*AP*GP*CP*C)-3') | | Authors: | Lohoefener, J, Steinke, N, Kay-Fedorov, P, Baruch, P, Nikulin, A, Tishchenko, S, Manstein, D.J, Fedorov, R. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The Activation Mechanism of 2'-5'-Oligoadenylate Synthetase Gives New Insights Into OAS/cGAS Triggers of Innate Immunity.
Structure, 23, 2015
|
|
4RWQ
 
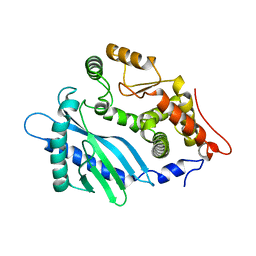 | | Crystal structure of the apo-state of porcine OAS1 | | Descriptor: | 2'-5'-oligoadenylate synthase 1 | | Authors: | Lohoefener, J, Steinke, N, Kay-Fedorov, P, Baruch, P, Nikulin, A, Tishchenko, S, Manstein, D.J, Fedorov, R. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The Activation Mechanism of 2'-5'-Oligoadenylate Synthetase Gives New Insights Into OAS/cGAS Triggers of Innate Immunity.
Structure, 23, 2015
|
|
4RWN
 
 | | Crystal structure of the pre-reactive state of porcine OAS1 | | Descriptor: | 2'-5'-oligoadenylate synthase 1, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, ... | | Authors: | Lohoefener, J, Steinke, N, Kay-Fedorov, P, Baruch, P, Nikulin, A, Tishchenko, S, Manstein, D.J, Fedorov, R. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Activation Mechanism of 2'-5'-Oligoadenylate Synthetase Gives New Insights Into OAS/cGAS Triggers of Innate Immunity.
Structure, 23, 2015
|
|
4RWO
 
 | | Crystal structure of the porcine OAS1 L149R mutant in complex with dsRNA and ApCpp in the AMP donor position | | Descriptor: | 2'-5'-oligoadenylate synthase 1, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, ... | | Authors: | Lohoefener, J, Steinke, N, Kay-Fedorov, P, Baruch, P, Nikulin, A, Tishchenko, S, Manstein, D.J, Fedorov, R. | | Deposit date: | 2014-12-05 | | Release date: | 2015-05-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Activation Mechanism of 2'-5'-Oligoadenylate Synthetase Gives New Insights Into OAS/cGAS Triggers of Innate Immunity.
Structure, 23, 2015
|
|
7TOC
 
 | | Crystal Structure of the Mitochondrial Ketol-acid Reductoisomerase IlvC from Candida auris | | Descriptor: | ACETIC ACID, Ketol-acid reductoisomerase, mitochondrial, ... | | Authors: | Kim, Y, Evdokimova, E, Di, R, Stogios, P, Savchenko, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2022-01-24 | | Release date: | 2022-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Crystal Structure of the Mitochondrial Ketol-acid Reductoisomerase IlvC from Candida auris
To Be Published
|
|
6ZX9
 
 | | Crystal structure of SIV Vpr,fused to T4 lysozyme, isolated from moustached monkey, bound to human DDB1 and human DCAF1 (amino acid residues 1046-1396) | | Descriptor: | DDB1- and CUL4-associated factor 1, DNA damage-binding protein 1, GLYCEROL, ... | | Authors: | Schwefel, D, Banchenko, S. | | Deposit date: | 2020-07-29 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.519729 Å) | | Cite: | Structural insights into Cullin4-RING ubiquitin ligase remodelling by Vpr from simian immunodeficiency viruses.
Plos Pathog., 17, 2021
|
|
6HD0
 
 | |
6YVG
 
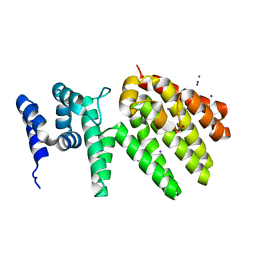 | | Crystal structure of MesI (Lpg2505) from Legionella pneumophila | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, MesI (Lpg2505) | | Authors: | Machtens, D.A, Willerding, J.M, Eschenburg, S, Reubold, T.F. | | Deposit date: | 2020-04-28 | | Release date: | 2020-06-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the metaeffector MesI (Lpg2505) from Legionella pneumophila.
Biochem.Biophys.Res.Commun., 527, 2020
|
|
6HD3
 
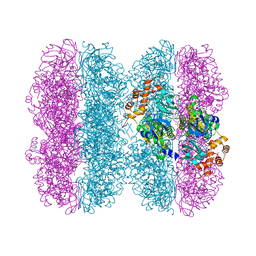 | | Common mode of remodeling AAA ATPases p97/CDC48 by their disassembly cofactors ASPL/PUX1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 48 homolog A, PHOSPHATE ION | | Authors: | Heinemann, U, Roske, Y, Banchenko, S, Arumughan, A, Petrovic, S. | | Deposit date: | 2018-08-17 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Common Mode of Remodeling AAA ATPases p97/CDC48 by Their Disassembling Cofactors ASPL/PUX1.
Structure, 27, 2019
|
|
