3RPT
 
 | |
7D9R
 
 | | Structure of huamn soluble guanylate cyclase in the riociguat and NO-bound state | | 分子名称: | Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1, MAGNESIUM ION, ... | | 著者 | Chen, L, Liu, R, Kang, Y. | | 登録日 | 2020-10-14 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.7 Å) | | 主引用文献 | Activation mechanism of human soluble guanylate cyclase by stimulators and activators.
Nat Commun, 12, 2021
|
|
7D9U
 
 | | Structure of human soluble guanylate cyclase in the cinciguat-bound activated state | | 分子名称: | 4-({(4-carboxybutyl)[2-(2-{[4-(2-phenylethyl)benzyl]oxy}phenyl)ethyl]amino}methyl)benzoic acid, Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1, ... | | 著者 | Chen, L, Liu, R, Kang, Y. | | 登録日 | 2020-10-14 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Activation mechanism of human soluble guanylate cyclase by stimulators and activators.
Nat Commun, 12, 2021
|
|
7D9S
 
 | | Structure of huamn soluble guanylate cyclase in the YC1 and NO-bound state | | 分子名称: | Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1, MAGNESIUM ION, ... | | 著者 | Chen, L, Liu, R, Kang, Y. | | 登録日 | 2020-10-14 | | 公開日 | 2021-08-11 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Activation mechanism of human soluble guanylate cyclase by stimulators and activators.
Nat Commun, 12, 2021
|
|
7D9T
 
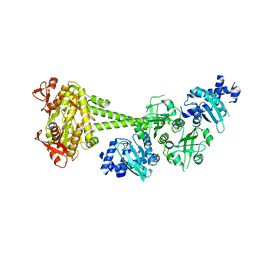 | |
6JPF
 
 | |
6JT2
 
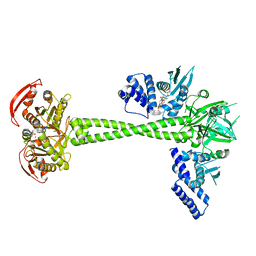 | | Structure of human soluble guanylate cyclase in the NO activated state | | 分子名称: | Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1, MAGNESIUM ION, ... | | 著者 | Chen, L, Kang, Y, Liu, R, Wu, J.-X. | | 登録日 | 2019-04-08 | | 公開日 | 2019-09-04 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Structural insights into the mechanism of human soluble guanylate cyclase.
Nature, 574, 2019
|
|
6L65
 
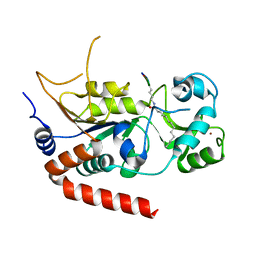 | |
6L66
 
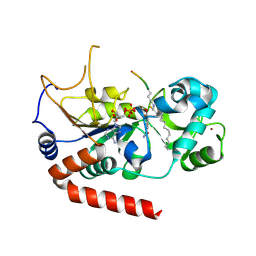 | |
6JT1
 
 | | Structure of human soluble guanylate cyclase in the heme oxidised state | | 分子名称: | Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Chen, L, Kang, Y, Liu, R, Wu, J.-X. | | 登録日 | 2019-04-08 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Structural insights into the mechanism of human soluble guanylate cyclase.
Nature, 574, 2019
|
|
6JB3
 
 | | Structure of SUR1 subunit bound with repaglinide | | 分子名称: | ATP-binding cassette sub-family C member 8 isoform X2, Digitonin, Repaglinide | | 著者 | Chen, L, Ding, D, Wang, M, Wu, J.-X, Kang, Y. | | 登録日 | 2019-01-25 | | 公開日 | 2019-05-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | ELECTRON MICROSCOPY (3.53 Å) | | 主引用文献 | The Structural Basis for the Binding of Repaglinide to the Pancreatic KATPChannel.
Cell Rep, 27, 2019
|
|
6JT0
 
 | | Structure of human soluble guanylate cyclase in the unliganded state | | 分子名称: | Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1, PROTOPORPHYRIN IX CONTAINING FE | | 著者 | Chen, L, Kang, Y, Liu, R, Wu, J.-X. | | 登録日 | 2019-04-08 | | 公開日 | 2019-08-28 | | 最終更新日 | 2024-03-27 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Structural insights into the mechanism of human soluble guanylate cyclase.
Nature, 574, 2019
|
|
6L72
 
 | | Sirtuin 2 demyristoylation native final product | | 分子名称: | NAD-dependent protein deacetylase sirtuin-2, ZINC ION, [(2S,3R,4R,5R)-5-[[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-2,4-bis(oxidanyl)oxolan-3-yl] tetradecanoate | | 著者 | Chen, L.F. | | 登録日 | 2019-10-30 | | 公開日 | 2021-03-31 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.501 Å) | | 主引用文献 | Sirtuin 2 protein with H3K18 myristoylated peptide
To Be Published
|
|
6L71
 
 | |
7VLB
 
 | |
7VVH
 
 | |
7VVD
 
 | |
7YEQ
 
 | |
7VUO
 
 | |
3R0M
 
 | | Crystal structure of anti-HIV llama VHH antibody A12 | | 分子名称: | Llama VHH A12, SULFATE ION | | 著者 | Chen, L, McLellan, J.S, Kwon, Y.D, Schmidt, S, Wu, X, Zhou, T, Yang, Y, Zhang, B, Forsman, A, Weiss, R.A, Verrips, T, Mascola, J, Kwong, P.D. | | 登録日 | 2011-03-08 | | 公開日 | 2012-03-14 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Single-Headed Immunoglobulins Efficiently Penetrate CD4-Binding Site and Effectively Neutralize HIV-1
To be Published
|
|
3RJQ
 
 | | Crystal structure of anti-HIV llama VHH antibody A12 in complex with C186 gp120 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C186 gp120, Llama VHH A12 | | 著者 | Chen, L, McLellan, J.S, Kwon, Y.D, Schmidt, S, Wu, X, Zhou, T, Yang, Y, Zhang, B, Forsman, A, Weiss, R.A, Verrips, T, Mascola, J, Kwong, P.D. | | 登録日 | 2011-04-15 | | 公開日 | 2012-05-02 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.601 Å) | | 主引用文献 | Crystal structure of anti-HIV A12 VHH of llama antibody in complex with C1086 gp120
To be Published
|
|
2HJM
 
 | |
7E16
 
 | |
7BR3
 
 | | Crystal structure of the protein 1 | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2R)-2,3-dihydroxypropyl dodecanoate, 4-[[(1R)-2-[5-(2-fluoranyl-3-methoxy-phenyl)-3-[[2-fluoranyl-6-(trifluoromethyl)phenyl]methyl]-4-methyl-2,6-bis(oxidanylidene)pyrimidin-1-yl]-1-phenyl-ethyl]amino]butanoic acid, ... | | 著者 | Cheng, L, Shao, Z. | | 登録日 | 2020-03-26 | | 公開日 | 2020-10-07 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.79 Å) | | 主引用文献 | Structure of the human gonadotropin-releasing hormone receptor GnRH1R reveals an unusual ligand binding mode.
Nat Commun, 11, 2020
|
|
7ECA
 
 | |
