6Y8O
 
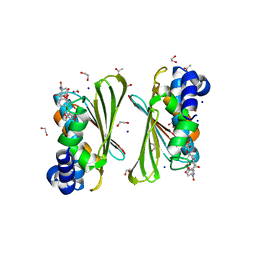 | | Mycobacterium smegmatis GyrB 22kDa ATPase sub-domain in complex with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA gyrase subunit B, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
6Y8L
 
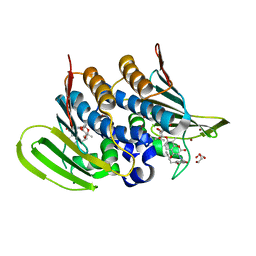 | | Mycobacterium thermoresistibile GyrB21 in complex with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, NOVOBIOCIN, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
4U6V
 
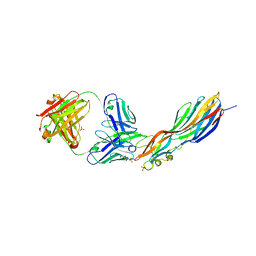 | | Mechanisms of Neutralization of a Human Anti-Alpha Toxin Antibody | | Descriptor: | Alpha-hemolysin, Fab, antigen binding fragment, ... | | Authors: | Oganesyan, V.Y, Peng, L, Damschroder, M.M, Cheng, L, Sadowska, A, Tkaczyk, C, Sellman, B, Wu, H, Dall'Acqua, W.F. | | Deposit date: | 2014-07-29 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Mechanisms of Neutralization of a Human Anti-alpha-toxin Antibody.
J.Biol.Chem., 289, 2014
|
|
6ZT5
 
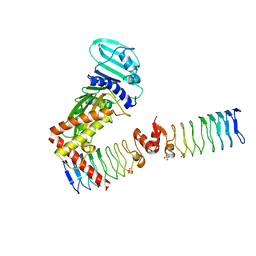 | | Complex between a homodimer of Mycobacterium smegmatis MfpA and a single copy of the N-terminal 47 kDa fragment of the Mycobacterium smegmatis DNA Gyrase B subunit | | Descriptor: | DNA gyrase subunit B, Pentapeptide repeat protein MfpA, SULFATE ION | | Authors: | Feng, L, Mundy, J.E.A, Stevenson, C.E.M, Mitchenall, L.A, Lawson, D.M, Mi, K, Maxwell, A. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The pentapeptide-repeat protein, MfpA, interacts with mycobacterial DNA gyrase as a DNA T-segment mimic.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6ZT4
 
 | | Pentapeptide repeat protein MfpA from Mycobacterium smegmatis | | Descriptor: | 1,2-ETHANEDIOL, Pentapeptide repeat protein MfpA | | Authors: | Feng, L, Mundy, J.E.A, Stevenson, C.E.M, Mitchenall, L.A, Lawson, D.M, Mi, K, Maxwell, A. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The pentapeptide-repeat protein, MfpA, interacts with mycobacterial DNA gyrase as a DNA T-segment mimic.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8JIL
 
 | | Cryo-EM structure of niacin bound ketone body receptor HCAR2-Gi signaling complex | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | Deposit date: | 2023-05-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
8JII
 
 | | Cryo-EM structure of compound 9n and niacin bound ketone body receptor HCAR2-Gi signaling complex | | Descriptor: | 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | Deposit date: | 2023-05-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
8JHY
 
 | | Cryo-EM structure of compound 9n bound ketone body receptor HCAR2-Gi signaling complex | | Descriptor: | 7-methyl-N-[(2R)-1-phenoxypropan-2-yl]-3-(4-propan-2-ylphenyl)pyrazolo[1,5-a]pyrimidine-6-carboxamide, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | Deposit date: | 2023-05-25 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
8JIM
 
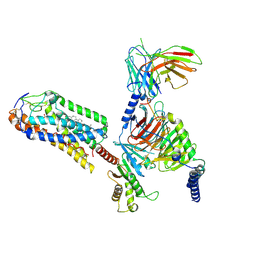 | | Cryo-EM structure of MMF bound ketone body receptor HCAR2-Gi signaling complex | | Descriptor: | (2Z)-4-methoxy-4-oxobut-2-enoic acid, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, C, Tian, X.W, Liu, Y, Cheng, L, Yan, W, Shao, Z.H. | | Deposit date: | 2023-05-26 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-27 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | Biased allosteric activation of ketone body receptor HCAR2 suppresses inflammation.
Mol.Cell, 83, 2023
|
|
5ZVS
 
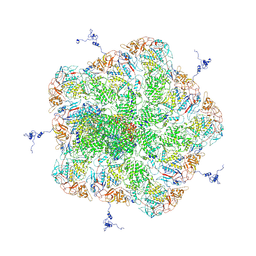 | |
6WE3
 
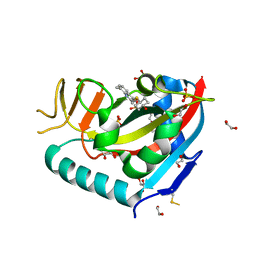 | | Human PARP14 (ARTD8), catalytic fragment in complex with compound 3 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(trans-4-hydroxycyclohexyl)sulfanyl]methyl}-8-methylquinazolin-4(3H)-one, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Swinger, K.S, Schenkel, L.B, Kuntz, K.W. | | Deposit date: | 2020-04-01 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A potent and selective PARP14 inhibitor decreases protumor macrophage gene expression and elicits inflammatory responses in tumor explants.
Cell Chem Biol, 28, 2021
|
|
6WE4
 
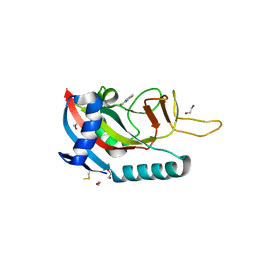 | | Human PARP14 (ARTD8), catalytic fragment in complex with compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-methyl-3,5,6,7-tetrahydro-4H-cyclopenta[4,5]thieno[2,3-d]pyrimidin-4-one, 8-methyl-2-{[(pyridin-4-yl)sulfanyl]methyl}quinazolin-4(3H)-one, ... | | Authors: | Swinger, K.S, Schenkel, L.B, Kuntz, K.W. | | Deposit date: | 2020-04-01 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A potent and selective PARP14 inhibitor decreases protumor macrophage gene expression and elicits inflammatory responses in tumor explants.
Cell Chem Biol, 28, 2021
|
|
1H9M
 
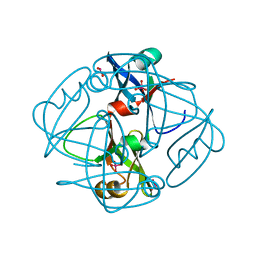 | | Two crystal structures of the cytoplasmic molybdate-binding protein ModG suggest a novel cooperative binding mechanism and provide insights into ligand-binding specificity. PEG-grown form with molybdate bound | | Descriptor: | MOLYBDATE ION, MOLYBDENUM-BINDING-PROTEIN | | Authors: | Delarbre, L, Stevenson, C.E.M, White, D.J, Mitchenall, L.A, Pau, R.N, Lawson, D.M. | | Deposit date: | 2001-03-13 | | Release date: | 2001-05-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Two Crystal Structures of the Cytoplasmic Molybdate-Binding Protein Modg Suggest a Novel Cooperative Binding Mechanism and Provide Insights Into Ligand-Binding Specificity
J.Mol.Biol., 308, 2001
|
|
5GON
 
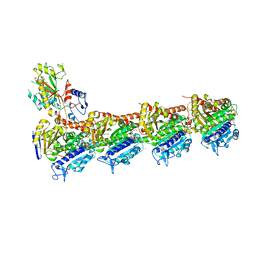 | | Structures of a beta-lactam bridged analogue in complex with tubulin | | Descriptor: | (3R,4R)-4-(4-methoxy-3-oxidanyl-phenyl)-3-methyl-1-(3,4,5-trimethoxyphenyl)azetidin-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Zhou, L, Liu, Y, Cheng, L, Wang, Y. | | Deposit date: | 2016-07-28 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Potent Antitumor Activities and Structure Basis of the Chiral beta-Lactam Bridged Analogue of Combretastatin A-4 Binding to Tubulin.
J. Med. Chem., 59, 2016
|
|
6L9I
 
 | |
4RN5
 
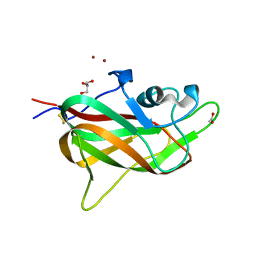 | | B1 domain of human Neuropilin-1 with acetate ion in a ligand-binding site | | Descriptor: | ACETATE ION, GLYCEROL, Neuropilin-1, ... | | Authors: | Allerston, C.K, Yelland, T.S, Jarvis, A, Jenkins, K, Winfield, N, Cheng, L, Jia, H, Zachary, I, Selwood, D.L, Djordjevic, S. | | Deposit date: | 2014-10-23 | | Release date: | 2015-10-28 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Conserved water molecules in a ligand-binding site of neuropilin-1
To be Published
|
|
6Y8N
 
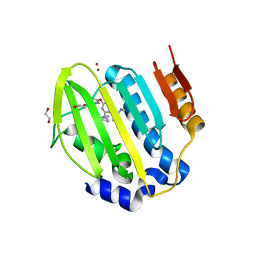 | | Mycobacterium thermoresistibile GyrB21 in complex with Redx03863 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(1~{S},5~{R})-6-azanyl-3-azabicyclo[3.1.0]hexan-3-yl]-6-fluoranyl-~{N}-methyl-2-(2-methylpyrimidin-5-yl)oxy-9~{H}-pyrimido[4,5-b]indol-8-amine, DNA gyrase subunit B, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
1H9J
 
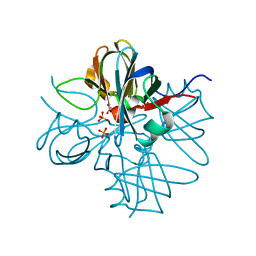 | | Two crystal structures of the cytoplasmic molybdate-binding protein ModG suggest a novel cooperative binding mechanism and provide insights into ligand-binding specificity. Phosphate-grown form with molybdate and phosphate bound | | Descriptor: | MOLYBDATE ION, MOLYBDENUM-BINDING-PROTEIN, PHOSPHATE ION | | Authors: | Delarbre, L, Stevenson, C.E.M, White, D.J, Mitchenall, L.A, Pau, R.N, Lawson, D.M. | | Deposit date: | 2001-03-13 | | Release date: | 2001-05-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two Crystal Structures of the Cytoplasmic Molybdate-Binding Protein Modg Suggest a Novel Cooperative Binding Mechanism and Provide Insights Into Ligand-Binding Specificity
J.Mol.Biol., 308, 2001
|
|
1H9K
 
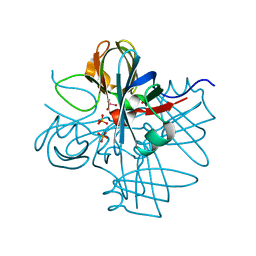 | | Two crystal structures of the cytoplasmic molybdate-binding protein ModG suggest a novel cooperative binding mechanism and provide insights into ligand-binding specificity. Phosphate-grown form with tungstate and phosphate bound | | Descriptor: | MOLYBDENUM-BINDING-PROTEIN, PHOSPHATE ION, TUNGSTATE(VI)ION | | Authors: | Delarbre, L, Stevenson, C.E.M, White, D.J, Mitchenall, L.A, Pau, R.N, Lawson, D.M. | | Deposit date: | 2001-03-13 | | Release date: | 2001-05-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Two Crystal Structures of the Cytoplasmic Molybdate-Binding Protein Modg Suggest a Novel Cooperative Binding Mechanism and Provide Insights Into Ligand-Binding Specificity
J.Mol.Biol., 308, 2001
|
|
4LO8
 
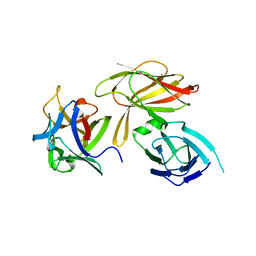 | | HA70(D3)-HA17 | | Descriptor: | HA-17, HA-70 | | Authors: | Lee, K, Gu, S, Jin, L, Le, T.T, Cheng, L.W, Strotmeier, J, Kruel, A.M, Yao, G, Perry, K, Rummel, A, Jin, R. | | Deposit date: | 2013-07-12 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Bimodular Botulinum Neurotoxin Complex Provides Insights into Its Oral Toxicity.
Plos Pathog., 9, 2013
|
|
2BM7
 
 | | The Structure of MfpA (Rv3361c, P3221 Crystal form). The Pentapeptide Repeat Protein from Mycobacterium tuberculosis Folds as A Right- handed Quadrilateral Beta-helix. | | Descriptor: | PENTAPEPTIDE REPEAT FAMILY PROTEIN | | Authors: | Hegde, S.S, Vetting, M.W, Roderick, S.L, Mitchenall, L.A, Maxwell, A, Takiff, H.E, Blanchard, J.S. | | Deposit date: | 2005-03-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Fluroquinolone Resistance Protein from Mycobacterium Tuberculosis that Mimics DNA
Science, 308, 2005
|
|
2BM6
 
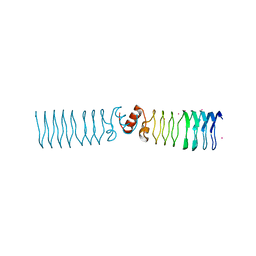 | | The Structure of MfpA (Rv3361c, C2221 Crystal form). The Pentapeptide Repeat Protein from Mycobacterium tuberculosis Folds as A Right- handed Quadrilateral Beta-helix. | | Descriptor: | CESIUM ION, PENTAPEPTIDE REPEAT FAMILY PROTEIN | | Authors: | Hegde, S.S, Vetting, M.W, Roderick, S.L, Mitchenall, L.A, Maxwell, A, Takiff, H.E, Blanchard, J.S. | | Deposit date: | 2005-03-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Fluroquinolone Resistance Protein from Mycobacterium Tuberculosis that Mimics DNA
Science, 308, 2005
|
|
2BM4
 
 | | The Structure of MfpA (Rv3361c, C2 Crystal form). The Pentapeptide Repeat Protein from Mycobacterium tuberculosis Folds as A Right- handed Quadrilateral Beta-helix. | | Descriptor: | PENTAPEPTIDE REPEAT FAMILY PROTEIN | | Authors: | Hegde, S.S, Vetting, M.W, Roderick, S.L, Mitchenall, L.A, Maxwell, A, Takiff, H.E, Blanchard, J.S. | | Deposit date: | 2005-03-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Fluroquinolone Resistance Protein from Mycobacterium Tuberculosis that Mimics DNA
Science, 308, 2005
|
|
2BM5
 
 | | The Structure of MfpA (Rv3361c, P21 Crystal form). The Pentapeptide Repeat Protein from Mycobacterium tuberculosis Folds as A Right- handed Quadrilateral Beta-helix. | | Descriptor: | PENTAPEPTIDE REPEAT FAMILY PROTEIN, SULFATE ION | | Authors: | Hegde, S.S, Vetting, M.W, Roderick, S.L, Mitchenall, L.A, Maxwell, A, Takiff, H.E, Blanchard, J.S. | | Deposit date: | 2005-03-09 | | Release date: | 2005-06-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Fluroquinolone Resistance Protein from Mycobacterium Tuberculosis that Mimics DNA
Science, 308, 2005
|
|
4LO3
 
 | | HA17-HA33-LacNac | | Descriptor: | HA-17, HA-33, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Lee, K, Gu, S, Jin, L, Le, T.T, Cheng, L.W, Strotmeier, J, Kruel, A.M, Yao, G, Perry, K, Rummel, A, Jin, R. | | Deposit date: | 2013-07-12 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.249 Å) | | Cite: | Structure of a Bimodular Botulinum Neurotoxin Complex Provides Insights into Its Oral Toxicity.
Plos Pathog., 9, 2013
|
|
