1QU1
 
 | | CRYSTAL STRUCTURE OF EHA2 (23-185) | | Descriptor: | PROTEIN (INFLUENZA RECOMBINANT HA2 CHAIN) | | Authors: | Chen, J, Skehel, J.J, Wiley, D.C. | | Deposit date: | 1999-07-05 | | Release date: | 2000-01-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA(2) subunit to form an N cap that terminates the triple-stranded coiled coil.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
7RDX
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - open class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RDZ
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - apo class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE3
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RDY
 
 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - engaged class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE0
 
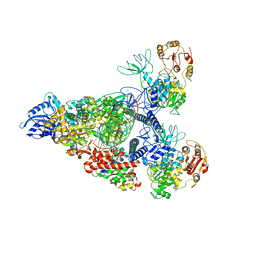 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - swiveled class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE2
 
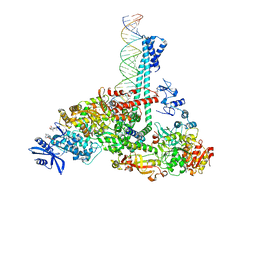 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(1)-RTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE1
 
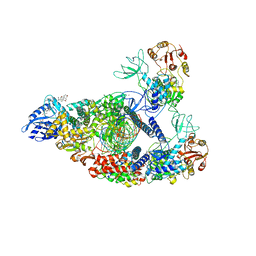 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC (composite) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7WZ5
 
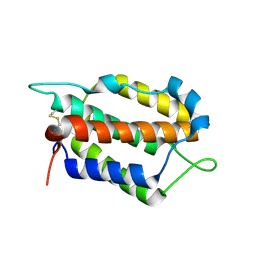 | | Larimichthys crocea IFNi | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interferon C | | Authors: | Chen, J.J. | | Deposit date: | 2022-02-17 | | Release date: | 2023-02-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Molecular and Structural Basis of Receptor Binding and Signaling of a Fish Type I IFN with Three Disulfide Bonds.
J Immunol., 209, 2022
|
|
8J60
 
 | | Structural and mechanistic insight into ribosomal ITS2 RNA processing by nuclease-kinase machinery | | Descriptor: | LAS1 protein, Polynucleotide 5'-hydroxyl-kinase GRC3 | | Authors: | Chen, J, Chen, H, Li, S, Lin, X, Hu, R, Zhang, K, Liu, L. | | Deposit date: | 2023-04-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural and mechanistic insights into ribosomal ITS2 RNA processing by nuclease-kinase machinery.
Elife, 12, 2024
|
|
8J5Y
 
 | | Structural and mechanistic insight into ribosomal ITS2 RNA processing by nuclease-kinase machinery | | Descriptor: | LAS1 isoform 1, Polynucleotide 5'-hydroxyl-kinase GRC3 | | Authors: | Chen, J, Chen, H, Li, S, Lin, X, Hu, R, Zhang, K, Liu, L. | | Deposit date: | 2023-04-24 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural and mechanistic insights into ribosomal ITS2 RNA processing by nuclease-kinase machinery.
Elife, 12, 2024
|
|
6EYO
 
 | | Structure of extended IgE-Fc in complex with two anti-IgE Fabs | | Descriptor: | 8D6 Fab heavy chain, 8D6 Fab light chain, Immunoglobulin heavy constant epsilon, ... | | Authors: | Chen, J.B, Ramadani, F, Pang, M.O.Y, Beavil, R.L, Holdom, M.D, Mitropoulou, A.N, Beavil, A.J, Gould, H.J, Chang, T.W, Sutton, B.J, McDonnell, J.M, Davies, A.M. | | Deposit date: | 2017-11-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody.
Sci Rep, 8, 2018
|
|
6EYN
 
 | | Structure of the 8D6 (anti-IgE) Fab | | Descriptor: | 1,2-ETHANEDIOL, 8D6 Fab heavy chain, 8D6 Fab light chain, ... | | Authors: | Chen, J.B, Ramadani, F, Pang, M.O.Y, Beavil, R.L, Holdom, M.D, Mitropoulou, A.N, Beavil, A.J, Gould, H.J, Chang, T.W, Sutton, B.J, McDonnell, J.M, Davies, A.M. | | Deposit date: | 2017-11-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for selective inhibition of immunoglobulin E-receptor interactions by an anti-IgE antibody.
Sci Rep, 8, 2018
|
|
4XUK
 
 | |
8IRQ
 
 | | Larimichthys crocea IFNd | | Descriptor: | Interferon d | | Authors: | Chen, J.J. | | Deposit date: | 2023-03-19 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of large yellow croaker IFNd at 1.49 Angstrom resolution.
To Be Published
|
|
4GY5
 
 | | Crystal structure of the tandem tudor domain and plant homeodomain of UHRF1 with Histone H3K9me3 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Peptide from Histone H3.3, ZINC ION | | Authors: | Cheng, J, Yang, Y, Fang, J, Xiao, J, Zhu, T, Chen, F, Wang, P, Xu, Y. | | Deposit date: | 2012-09-05 | | Release date: | 2012-11-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.956 Å) | | Cite: | Structural insight into coordinated recognition of trimethylated histone H3 lysine 9 (H3K9me3) by the plant homeodomain (PHD) and tandem tudor domain (TTD) of UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) protein
J.Biol.Chem., 288, 2013
|
|
6PSU
 
 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPi2) with TraR and rpsT P2 promoter | | Descriptor: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
6PSR
 
 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPi1) with TraR and rpsT P2 promoter | | Descriptor: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
6PSW
 
 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPo) with TraR and rpsT P2 promoter | | Descriptor: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
6PSS
 
 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPi1.5a) with TraR and mutant rpsT P2 promoter | | Descriptor: | DNA (85-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
6PSQ
 
 | | Escherichia coli RNA polymerase closed complex (TRPc) with TraR and rpsT P2 promoter | | Descriptor: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
6PST
 
 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TRPi1.5b) with TraR and mutant rpsT P2 promoter | | Descriptor: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
6PSV
 
 | | Escherichia coli RNA polymerase promoter unwinding intermediate (TpreRPo) with TraR and rpsT P2 promoter | | Descriptor: | CHAPSO, DNA (85-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-07-13 | | Release date: | 2020-03-25 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Stepwise Promoter Melting by Bacterial RNA Polymerase.
Mol.Cell, 78, 2020
|
|
6OUL
 
 | | Cryo-EM structure of Escherichia coli RNAP polymerase bound to rpsTP2 promoter DNA | | Descriptor: | CHAPSO, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-05-04 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | E. coliTraR allosterically regulates transcription initiation by altering RNA polymerase conformation.
Elife, 8, 2019
|
|
6P1K
 
 | | Cryo-EM structure of Escherichia coli sigma70 bound RNAP polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Chen, J, Chiu, C.E, Campbell, E.A, Darst, S.A. | | Deposit date: | 2019-05-20 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | E. coliTraR allosterically regulates transcription initiation by altering RNA polymerase conformation.
Elife, 8, 2019
|
|
