6RZ9
 
 | | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2770372 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S})-8-[[4-[4-(5-fluoranyl-2-methyl-phenyl)butoxy]phenyl]carbonylamino]-4-(4-oxidanyl-4-oxidanylidene-butyl)-2,3-dih ydro-1,4-benzoxazine-2-carboxylic acid, CHOLESTEROL, ... | | Authors: | Gusach, A, Luginina, A, Marin, E, Brouillette, R.L, Besserer-Offroy, E, Longpre, J.M, Ishchenko, A, Popov, P, Fujimoto, T, Maruyama, T, Stauch, B, Ergasheva, M, Romanovskaya, D, Stepko, A, Kovalev, K, Shevtsov, M, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors.
Nat Commun, 10, 2019
|
|
6BNF
 
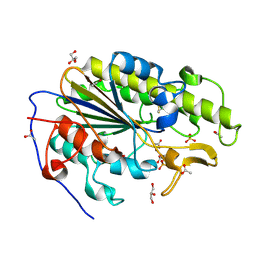 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, mono-zinc complex | | Descriptor: | ACETATE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Di Leo, R, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Substrate recognition by a colistin resistance enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 2018
|
|
4JG9
 
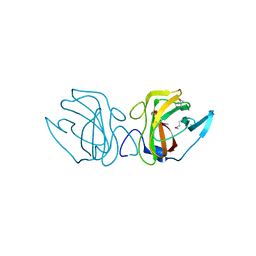 | | X-ray Crystal Structure of a Putative Lipoprotein from Bacillus anthracis | | Descriptor: | Lipoprotein | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-02-28 | | Release date: | 2013-03-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.425 Å) | | Cite: | X-ray Crystal Structure of a Putative Lipoprotein from Bacillus anthracis
To be Published
|
|
3L44
 
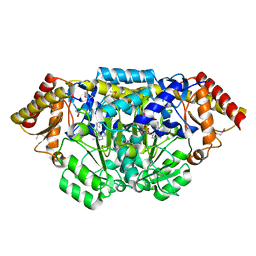 | | Crystal structure of Bacillus anthracis HemL-1, glutamate semialdehyde aminotransferase | | Descriptor: | Glutamate-1-semialdehyde 2,1-aminomutase 1 | | Authors: | Anderson, S.M, Wawrzak, Z, Gordon, E, Hasseman, J, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Bacillus anthracis HemL-1, glutamate semialdehyde aminotransferase
TO BE PUBLISHED
|
|
3I0U
 
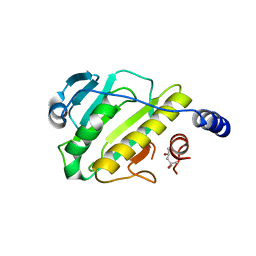 | | Structure of the type III effector/phosphothreonine lyase OspF from Shigella flexneri | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Phosphothreonine lyase ospF | | Authors: | Singer, A.U, Skarina, T, Nocek, B, Gordon, R, Lam, R, Kagan, O, Edwards, A.M, Joachimiak, A, Chirgadze, N.Y, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-06-25 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the type III effector/phosphothreonine lyase OspF from Shigella flexneri
TO BE PUBLISHED
|
|
6BND
 
 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, Thr315Ala mutant mono-zinc and phosphoethanolamine complex | | Descriptor: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, POLYETHYLENE GLYCOL (N=34), Phosphoethanolamine transferase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Substrate Recognition by a Colistin Resistance Enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 13, 2018
|
|
6BNE
 
 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, phosphate-bound complex | | Descriptor: | ACETATE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Di Leo, R, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Substrate recognition by a colistin resistance enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 2018
|
|
3KEB
 
 | | Thiol peroxidase from Chromobacterium violaceum | | Descriptor: | CHLORIDE ION, Probable thiol peroxidase, SULFATE ION | | Authors: | Osipiuk, J, Kagan, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-10-25 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of thiol peroxidase from Chromobacterium violaceum
To be Published
|
|
6RZ6
 
 | | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2570366 (C2221 space group) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S})-8-[[4-[4-(2-chloranyl-5-fluoranyl-phenyl)butoxy]phenyl]carbonylamino]-4-(4-oxidanyl-4-oxidanylidene-butyl)-2,3- dihydro-1,4-benzoxazine-2-carboxylic acid, CHOLESTEROL, ... | | Authors: | Gusach, A, Luginina, A, Marin, E, Brouillette, R.L, Besserer-Offroy, E, Longpre, J.M, Ishchenko, A, Popov, P, Fujimoto, T, Maruyama, T, Stauch, B, Ergasheva, M, Romanovskaya, D, Stepko, A, Kovalev, K, Shevtsov, M, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors.
Nat Commun, 10, 2019
|
|
6CD7
 
 | | Crystal structure of APH(2")-IVa in complex with plazomicin | | Descriptor: | (2S)-4-amino-N-[(1R,2S,3S,4R,5S)-5-amino-4-{[(2S,3R)-3-amino-6-{[(2-hydroxyethyl)amino]methyl}-3,4-dihydro-2H-pyran-2-y l]oxy}-2-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-3-hydroxycyclohexyl]-2-hydroxybutanamide, APH(2'')-Id, CHLORIDE ION | | Authors: | Stogios, P.J, Evdokimova, E, Dong, A, Di Leo, R, Savchenko, A, Satchell, K.J, Joachimiak, J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-02-08 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Plazomicin Retains Antibiotic Activity against Most Aminoglycoside Modifying Enzymes.
ACS Infect Dis, 4, 2018
|
|
4H5G
 
 | | Crystal structure of an amino acid ABC transporter substrate-binding protein from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine, form 2 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ARGININE, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Minasov, G, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-18 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of an amino acid ABC transporter substrate-binding protein from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine, form 2
TO BE PUBLISHED
|
|
4H5F
 
 | | Crystal structure of an amino acid ABC transporter substrate-binding protein from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine, form 1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, ARGININE, ... | | Authors: | Stogios, P.J, Wawrzak, Z, Kudritska, M, Minasov, G, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-09-18 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an amino acid ABC transporter substrate-binding protein from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine, form 1
TO BE PUBLISHED
|
|
5K2B
 
 | | 2.5 angstrom A2a adenosine receptor structure with MR phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
4JID
 
 | | Crystal structure of BaLdcB / VanY-like L,D-carboxypeptidase Zinc(II)-free | | Descriptor: | CHLORIDE ION, D-alanyl-D-alanine carboxypeptidase family protein | | Authors: | Minasov, G, Wawrzak, Z, Onopriyenko, O, Skarina, T, Shatsman, S, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-05 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the LdcB LD-Carboxypeptidase Reveals the Molecular Basis of Peptidoglycan Recognition.
Structure, 22, 2014
|
|
5DPO
 
 | |
5K2D
 
 | | 1.9A angstrom A2a adenosine receptor structure with MR phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
3I1J
 
 | | Structure of a putative short chain dehydrogenase from Pseudomonas syringae | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Singer, A.U, Evdokimova, E, Kudritska, M, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-06-26 | | Release date: | 2009-07-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a putative short chain dehydrogenase from Pseudomonas syringae
To be Published
|
|
5BY0
 
 | | Crystal structure of magnesium-bound Duf89 protein Saccharomyces cerevisiae | | Descriptor: | MAGNESIUM ION, Protein-glutamate O-methyltransferase | | Authors: | Nocek, B, Cuff, M, Cui, H, Xu, X, Savchenko, A, Joachimiak, A, Yakunin, A. | | Deposit date: | 2015-06-09 | | Release date: | 2015-07-29 | | Last modified: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of magnesium-bound Duf89 protein Saccharomyces cerevisiae
To Be Published
|
|
5K2C
 
 | | 1.9 angstrom A2a adenosine receptor structure with sulfur SAD phasing and phase extension using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
5K2A
 
 | | 2.5 angstrom A2a adenosine receptor structure with sulfur SAD phasing using XFEL data | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 chimera, ... | | Authors: | Batyuk, A, Galli, L, Ishchenko, A, Han, G.W, Gati, C, Popov, P, Lee, M.-Y, Stauch, B, White, T.A, Barty, A, Aquila, A, Hunter, M.S, Liang, M, Boutet, S, Pu, M, Liu, Z.-J, Nelson, G, James, D, Li, C, Zhao, Y, Spence, J.C.H, Liu, W, Fromme, P, Katritch, V, Weierstall, U, Stevens, R.C, Cherezov, V, GPCR Network (GPCR) | | Deposit date: | 2016-05-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Native phasing of x-ray free-electron laser data for a G protein-coupled receptor.
Sci Adv, 2, 2016
|
|
5D8M
 
 | | Crystal structure of the metagenomic carboxyl esterase MGS0156 | | Descriptor: | Metagenomic carboxyl esterase MGS0156 | | Authors: | Cui, H, Nocek, B, Tchigvintsev, A, Popovic, A, Savchenko, A, Joachimiak, A, Yakunin, A. | | Deposit date: | 2015-08-17 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of esterase (MGS0156)
To Be Published
|
|
3KQF
 
 | | 1.8 Angstrom Resolution Crystal Structure of Enoyl-CoA Hydratase from Bacillus anthracis. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Enoyl-CoA hydratase/isomerase family protein | | Authors: | Minasov, G, Halavaty, A, Wawrzak, Z, Skarina, T, Onopriyenko, O, Papazisi, L, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-11-17 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Enoyl-CoA Hydratase from Bacillus anthracis.
TO BE PUBLISHED
|
|
3H3M
 
 | | Crystal structure of flagellar protein FliT from Bordetella bronchiseptica | | Descriptor: | Flagellar protein FliT, UNKNOWN | | Authors: | Shumilin, I.A, Wang, S, Chruszcz, M, Xu, X, Le, B, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-04-16 | | Release date: | 2009-04-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of flagellar protein FliT from Bordetella bronchiseptica
To be Published
|
|
3GL5
 
 | | Crystal structure of probable DsbA oxidoreductase SCO1869 from Streptomyces coelicolor | | Descriptor: | ACETATE ION, Putative DsbA oxidoreductase SCO1869, SODIUM ION | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-03-11 | | Release date: | 2009-03-31 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of probable DsbA oxidoreductase SCO1869 from Streptomyces coelicolor
To be Published
|
|
3IAH
 
 | | Crystal Structure of Short Chain Dehydrogenase (yciK) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 in Complex with NADP and Acetate. | | Descriptor: | ACETATE ION, CHLORIDE ION, ETHANOL, ... | | Authors: | Minasov, G, Halavaty, A, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-07-14 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal Structure of Short Chain Dehydrogenase (yciK) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 in Complex with NADP and Acetate.
TO BE PUBLISHED
|
|
