3HFY
 
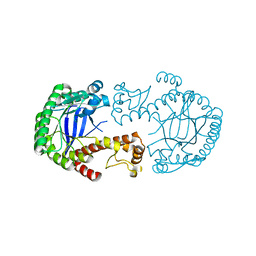 | | Mutant of tRNA-guanine transglycosylase (K52M) | | Descriptor: | Queuine tRNA-ribosyltransferase, ZINC ION | | Authors: | Ritschel, T, Klebe, G. | | Deposit date: | 2009-05-13 | | Release date: | 2009-08-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An Integrative Approach Combining Noncovalent Mass Spectrometry, Enzyme Kinetics and X-ray Crystallography to Decipher Tgt Protein-Protein and Protein-RNA Interaction
J.Mol.Biol., 393, 2009
|
|
3EOS
 
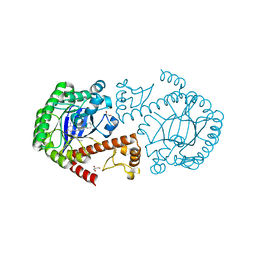 | | tRNA-guanine transglycosylase in complex with 6-amino-4-{2-[(cyclohexylmethyl)amino]ethyl}-2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-4-{2-[(cyclohexylmethyl)amino]ethyl}-2-(methylamino)-1,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Ritschel, T, Heine, A, Klebe, G. | | Deposit date: | 2008-09-29 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | How to Replace the Residual Solvation Shell of Polar Active Site Residues to Achieve Nanomolar Inhibition of tRNA-Guanine Transglycosylase
Chemmedchem, 4, 2009
|
|
2HN7
 
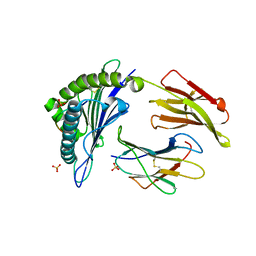 | | HLA-A*1101 in complex with HBV peptide homologue | | Descriptor: | Beta-2-microglobulin, DNA polymerase PEPTIDE HOMOLOGUE, HLA class I histocompatibility antigen, ... | | Authors: | Blicher, T. | | Deposit date: | 2006-07-12 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of HLA-A*1101 in complex with a hepatitis B peptide homologue.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
5WIU
 
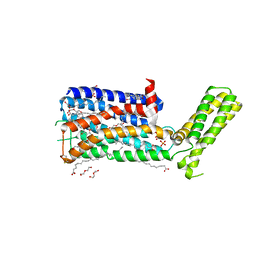 | | Structure of the human D4 Dopamine receptor in complex with Nemonapride | | Descriptor: | D(4) dopamine receptor, soluble cytochrome b562 chimera, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wacker, D, Wang, S, Levit, A, Che, T, Betz, R.M, McCorvy, J.D, Venkatakrishnan, A.J, Huang, X.-P, Dror, R.O, Shoichet, B.K, Roth, B.L. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | D4 dopamine receptor high-resolution structures enable the discovery of selective agonists.
Science, 358, 2017
|
|
5WIV
 
 | | Structure of the sodium-bound human D4 Dopamine receptor in complex with Nemonapride | | Descriptor: | D(4) dopamine receptor, soluble cytochrome b562 chimera, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wacker, D, Wang, S, Levit, A, Che, T, Betz, R.M, McCorvy, J.D, Venkatakrishnan, A.J, Huang, X.-P, Dror, R.O, Shoichet, B.K, Roth, B.L. | | Deposit date: | 2017-07-20 | | Release date: | 2017-10-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | D4 dopamine receptor high-resolution structures enable the discovery of selective agonists.
Science, 358, 2017
|
|
5J4K
 
 | | Structure of humanised RadA-mutant humRadA22F in complex with 1-Indane-6-carboxylic acid | | Descriptor: | 2,3-dihydro-1H-indene-2-carboxylic acid, CALCIUM ION, DNA repair and recombination protein RadA, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.346 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5JEC
 
 | | Apo-structure of humanised RadA-mutant humRadA33F | | Descriptor: | CHLORIDE ION, DNA repair and recombination protein RadA, SULFATE ION | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5JED
 
 | | Apo-structure of humanised RadA-mutant humRadA28 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5J4H
 
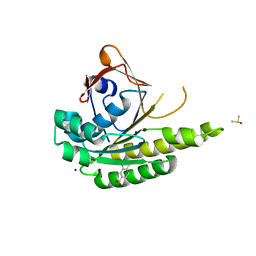 | | Structure of humanised RadA-mutant humRadA22F in complex with indole-6-carboxylic acid | | Descriptor: | 1H-indole-6-carboxylic acid, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
8DZP
 
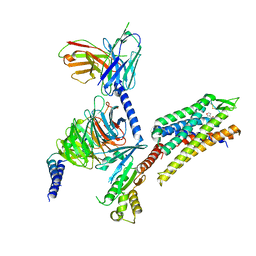 | | momSalB bound Kappa Opioid Receptor in complex with Gi1 | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Fay, J.F, Che, T. | | Deposit date: | 2022-08-08 | | Release date: | 2023-05-03 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Ligand and G-protein selectivity in the kappa-opioid receptor.
Nature, 617, 2023
|
|
8DZQ
 
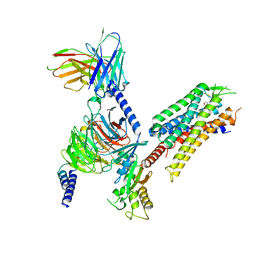 | | momSalB bound Kappa Opioid Receptor in complex with GoA | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(o) subunit alpha, ... | | Authors: | Fay, J.F, Che, T. | | Deposit date: | 2022-08-08 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.82 Å) | | Cite: | Ligand and G-protein selectivity in the kappa-opioid receptor.
Nature, 617, 2023
|
|
8DZR
 
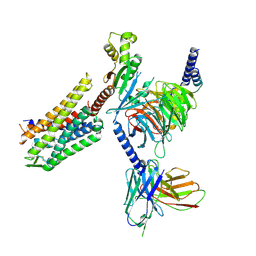 | | GR89,696 bound Kappa Opioid Receptor in complex with gustducin | | Descriptor: | G alpha gustducin protein, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Fay, J.F, Che, T. | | Deposit date: | 2022-08-08 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.61 Å) | | Cite: | Ligand and G-protein selectivity in the kappa-opioid receptor.
Nature, 617, 2023
|
|
8DZS
 
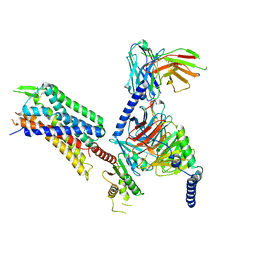 | | GR89,696 bound Kappa Opioid Receptor in complex with Gz | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(z) subunit alpha, ... | | Authors: | Fay, J.F, Che, T. | | Deposit date: | 2022-08-08 | | Release date: | 2023-05-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Ligand and G-protein selectivity in the kappa-opioid receptor.
Nature, 617, 2023
|
|
8FEG
 
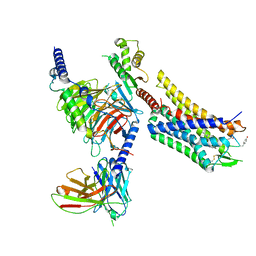 | | CryoEM structure of Kappa Opioid Receptor bound to a semi-peptide and Gi1 | | Descriptor: | ACE-TYR-ALA-DTY-THR-THR-CYS-THR-DPN-XT9, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Fay, J.F, Che, T. | | Deposit date: | 2022-12-06 | | Release date: | 2023-12-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Design and structural validation of peptide-drug conjugate ligands of the kappa-opioid receptor.
Nat Commun, 14, 2023
|
|
8D1P
 
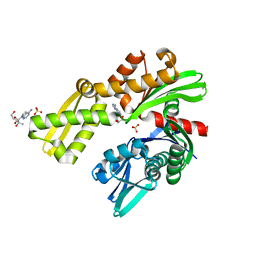 | | Crystal structure of Plasmodium falciparum GRP78-NBD in complex with 7-Deaza-2'-C-methyladenosine | | Descriptor: | 7-(2-C-methyl-beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine, Chaperone DnaK, SULFATE ION | | Authors: | Mrozek, A, Chen, Y, Antoshchenko, T, Park, H.W. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A non-traditional crystal-based compound screening method targeting the ATP binding site of Plasmodium falciparum GRP78 for identification of novel nucleoside analogues.
Front Mol Biosci, 9, 2022
|
|
8D1S
 
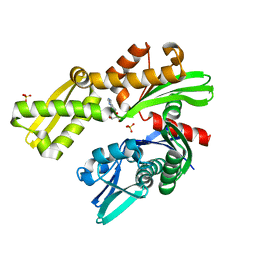 | | Crystal structure of Plasmodium falciparum GRP78 in complex with Toyocamycin | | Descriptor: | 4-amino-7-(beta-D-ribofuranosyl)-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Chaperone DnaK, SULFATE ION | | Authors: | Mrozek, A, Chen, Y, Antoshchenko, T, Park, H.W. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A non-traditional crystal-based compound screening method targeting the ATP binding site of Plasmodium falciparum GRP78 for identification of novel nucleoside analogues.
Front Mol Biosci, 9, 2022
|
|
8D1Q
 
 | | Crystal structure of Plasmodium falciparum GRP78-NBD in complex with 8-Aminoadenosine | | Descriptor: | (2R,3R,4S,5R)-2-(6,8-diaminopurin-9-yl)-5-(hydroxymethyl)oxolane-3,4-diol, Chaperone DnaK, SULFATE ION | | Authors: | Mrozek, A, Chen, Y, Antoshchenko, T, Park, H.W. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A non-traditional crystal-based compound screening method targeting the ATP binding site of Plasmodium falciparum GRP78 for identification of novel nucleoside analogues.
Front Mol Biosci, 9, 2022
|
|
6BQG
 
 | | Crystal structure of 5-HT2C in complex with ergotamine | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562, Ergotamine | | Authors: | Peng, Y, McCorvy, J.D, Harpsoe, K, Lansu, K, Yuan, S, Popov, P, Qu, L, Pu, M, Che, T, Nikolajse, L.F, Huang, X.P, Wu, Y, Shen, L, Bjorn-Yoshimoto, W.E, Ding, K, Wacker, D, Han, G.W, Cheng, J, Katritch, V, Jensen, A.A, Hanson, M.A, Zhao, S, Gloriam, D.E, Roth, B.L, Stevens, R.C, Liu, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology.
Cell, 172, 2018
|
|
6BQH
 
 | | Crystal structure of 5-HT2C in complex with ritanserin | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562, 6-(2-{4-[bis(4-fluorophenyl)methylidene]piperidin-1-yl}ethyl)-7-methyl-5H-[1,3]thiazolo[3,2-a]pyrimidin-5-one, ... | | Authors: | Peng, Y, McCorvy, J.D, Harpsoe, K, Lansu, K, Yuan, S, Popov, P, Qu, L, Pu, M, Che, T, Nikolajse, L.F, Huang, X.P, Wu, Y, Shen, L, Bjorn-Yoshimoto, W.E, Ding, K, Wacker, D, Han, G.W, Cheng, J, Katritch, V, Jensen, A.A, Hanson, M.A, Zhao, S, Gloriam, D.E, Roth, B.L, Stevens, R.C, Liu, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology.
Cell, 172, 2018
|
|
7YIT
 
 | |
8D1W
 
 | | Crystal structure of Plasmodium falciparum GRP78 in complex with (2R,3R,4S,5R)-2-(6-amino-8-((2-chlorobenzyl)amino)-9H-purin-9-yl)-5-(hydroxymethyl)tetrahydrofuran-3,4-diol | | Descriptor: | 8-{[(2-chlorophenyl)methyl]amino}adenosine, Chaperone DnaK, SULFATE ION | | Authors: | Mrozek, A, Chen, Y, Antoshchenko, T, Park, H.W, Smil, D, Zepeda, C.A.V. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting Plasmodium falciparum GRP78: nucleoside analogues as agents against the malaria chaperone
Frontiers in Molecular Bioscience, 2022
|
|
9BEQ
 
 | | Crystal structure of pregnane X receptor ligand binding domain complexed with AP1867 | | Descriptor: | Nuclear receptor subfamily 1 group I member 2, {3-[3-(3,4-DIMETHOXY-PHENYL)-1-(1-{1-[2-(3,4,5-TRIMETHOXY-PHENYL)-BUTYRYL]-PIPERIDIN-2YL}-VINYLOXY)-PROPYL]-PHENOXY}-ACETIC ACID | | Authors: | Huber, A.D, Poudel, S, Miller, D.J, Chen, T. | | Deposit date: | 2024-04-16 | | Release date: | 2024-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | PROTAC-mediated activation, rather than degradation, of a nuclear receptor reveals complex ligand-receptor interaction network.
Structure, 2024
|
|
4MS2
 
 | | Structural basis of Ca2+ selectivity of a voltage-gated calcium channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Martinez, G.Q, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-18 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
4MTF
 
 | | Structural Basis of Ca2+ Selectivity of a Voltage-gated Calcium Channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Martinez, G.Q, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-19 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
4MTG
 
 | | Structural Basis for Ca2+ Selectivity of a Voltage-gated Calcium Channel | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, Ion transport protein | | Authors: | Tang, L, Gamal El-Din, T.M, Payandeh, J, Martinez, G.Q, Heard, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2013-09-19 | | Release date: | 2013-11-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.296 Å) | | Cite: | Structural basis for Ca2+ selectivity of a voltage-gated calcium channel.
Nature, 505, 2014
|
|
