3B61
 
 | |
3B5D
 
 | |
3B62
 
 | |
4LSG
 
 | |
4XWK
 
 | | P-glycoprotein co-crystallized with BDE-100 | | Descriptor: | 2,4-dibromophenyl 2,4,6-tribromophenyl ether, Multidrug resistance protein 1A | | Authors: | McGrath, A.P, Rees, S.D, Chang, G. | | Deposit date: | 2015-01-28 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Global marine pollutants inhibit P-glycoprotein: Environmental levels, inhibitory effects, and cocrystal structure.
Sci Adv, 2, 2016
|
|
6MRX
 
 | | Sialidase26 apo | | Descriptor: | Sialidase26 | | Authors: | Zaramela, L.S, Martino, C, Alisson-Silva, F, Rees, S.D, Diaz, S.L, Chuzel, L, Ganatra, M.B, Taron, C.H, Zuniga, C, Chang, G, Varki, A, Zengler, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
6MYV
 
 | | Sialidase26 co-crystallized with DANA-Gc | | Descriptor: | 2,6-anhydro-3,5-dideoxy-5-[(hydroxyacetyl)amino]-D-glycero-L-altro-non-2-enonic acid, Sialidase26 | | Authors: | Zaramela, L.S, Martino, C, Alisson-Silva, F, Rees, S.D, Diaz, S.L, Chuzel, L, Ganatra, M.B, Taron, C.H, Zuniga, C, Huang, J, Siegel, D, Chang, G, Varki, A, Zengler, K. | | Deposit date: | 2018-11-02 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
6MRV
 
 | | Sialidase26 co-crystallized with DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, Sialidase26 | | Authors: | Zaramela, L.S, Martino, C, Alisson-Silva, F, Rees, S.D, Diaz, S.L, Chuzel, L, Ganatra, M.B, Taron, C.H, Zuniga, C, Chang, G, Varki, A, Zengler, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
6MHF
 
 | | Galphai3 co-crystallized with GIV/Girdin | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, Girdin, ... | | Authors: | Rees, S.D, Kalogriopoulos, N.A, Ngo, T, Kopcho, N, Ilatovskiy, A, Sun, N, Komives, E, Chang, G, Ghosh, P, Kufareva, I. | | Deposit date: | 2018-09-17 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for GPCR-independent activation of heterotrimeric Gi proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6MNJ
 
 | | Hadza microbial sialidase Hz136 | | Descriptor: | Hz136 | | Authors: | Rees, S.D, Zaramela, L, Zengler, K, Chang, G. | | Deposit date: | 2018-10-01 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
6MHE
 
 | | Galphai3 co-crystallized with KB752 | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(k) subunit alpha, ... | | Authors: | Rees, S.D, Kalogriopoulos, N.A, Ngo, T, Kopcho, N, Ilatovskiy, A, Sun, N, Komives, E, Chang, G, Ghosh, P, Kufareva, I. | | Deposit date: | 2018-09-17 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for GPCR-independent activation of heterotrimeric Gi proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7MHU
 
 | | Sialidase24 apo | | Descriptor: | Exo-alpha-sialidase | | Authors: | Rees, S.D, Chang, G.A. | | Deposit date: | 2021-04-15 | | Release date: | 2022-02-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computational models in the service of X-ray and cryo-electron microscopy structure determination.
Proteins, 89, 2021
|
|
6OCE
 
 | | Structure of the rice hyperosmolality-gated ion channel OSCA1.2 | | Descriptor: | stress-gated cation channel 1.2 | | Authors: | Maity, K, Heumann, J.M, McGrath, A.P, Chang, G, Stowell, M.H. | | Deposit date: | 2019-03-23 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structure of OSCA1.2 fromOryza sativaelucidates the mechanical basis of potential membrane hyperosmolality gating.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3B5W
 
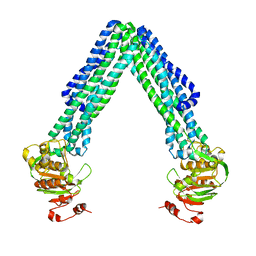 | | Crystal Structure of Eschericia coli MsbA | | Descriptor: | Lipid A export ATP-binding/permease protein msbA | | Authors: | Ward, A, Reyes, C.L, Yu, J, Roth, C.B, Chang, G. | | Deposit date: | 2007-10-26 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (5.3 Å) | | Cite: | Flexibility in the ABC transporter MsbA: Alternating access with a twist.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OOE
 
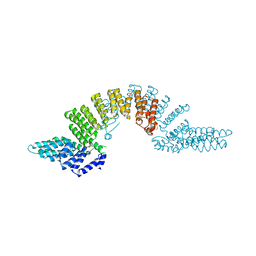 | | Crystal structure of HAT domain of murine CstF-77 | | Descriptor: | Cleavage stimulation factor 77 kDa subunit | | Authors: | Bai, Y, Auperin, T.C, Chou, C.-Y, Chang, G.-G, Manley, J.L, Tong, L. | | Deposit date: | 2007-01-25 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of Murine CstF-77: Dimeric Association and Implications for Polyadenylation of mRNA Precursors.
Mol.Cell, 25, 2007
|
|
2OND
 
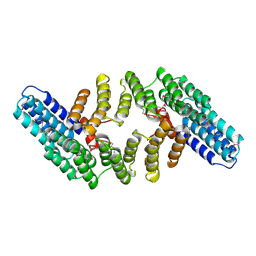 | | Crystal Structure of the HAT-C domain of murine CstF-77 | | Descriptor: | Cleavage stimulation factor 77 kDa subunit | | Authors: | Bai, Y, Auperin, T.C, Chou, C.-Y, Chang, G.-G, Manley, J.L, Tong, L. | | Deposit date: | 2007-01-23 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Murine CstF-77: Dimeric Association and Implications for Polyadenylation of mRNA Precursors.
Mol.Cell, 25, 2007
|
|
1PJL
 
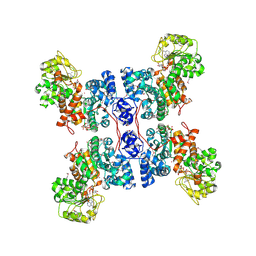 | | Crystal structure of human m-NAD-ME in ternary complex with NAD and Lu3+ | | Descriptor: | LUTETIUM (III) ION, NAD-dependent malic enzyme, mitochondrial, ... | | Authors: | Yang, Z, Batra, R, Floyd, D.L, Hung, H.-C, Chang, G.-G, Tong, L. | | Deposit date: | 2003-06-03 | | Release date: | 2003-06-17 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Potent and competitive inhibition of malic enzymes by lanthanide ions
Biochem.Biophys.Res.Commun., 274, 2000
|
|
3B5Z
 
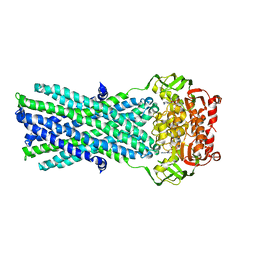 | | Crystal Structure of MsbA from Salmonella typhimurium with ADP Vanadate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lipid A export ATP-binding/permease protein msbA, VANADATE ION | | Authors: | Ward, A, Reyes, C.L, Yu, J, Roth, C.B, Chang, G. | | Deposit date: | 2007-10-26 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Flexibility in the ABC transporter MsbA: Alternating access with a twist.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3B60
 
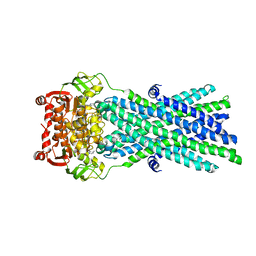 | | Crystal Structure of MsbA from Salmonella typhimurium with AMPPNP, higher resolution form | | Descriptor: | Lipid A export ATP-binding/permease protein msbA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Ward, A, Reyes, C.L, Yu, J, Roth, C.B, Chang, G. | | Deposit date: | 2007-10-26 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Flexibility in the ABC transporter MsbA: Alternating access with a twist.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3B5Y
 
 | | Crystal Structure of MsbA from Salmonella typhimurium with AMPPNP | | Descriptor: | Lipid A export ATP-binding/permease protein msbA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Ward, A, Reyes, C.L, Yu, J, Roth, C.B, Chang, G. | | Deposit date: | 2007-10-26 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Flexibility in the ABC transporter MsbA: Alternating access with a twist.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1LBG
 
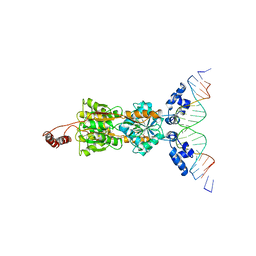 | | LACTOSE OPERON REPRESSOR BOUND TO 21-BASE PAIR SYMMETRIC OPERATOR DNA, ALPHA CARBONS ONLY | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*T)-3'), PROTEIN (LACTOSE OPERON REPRESSOR) | | Authors: | Lewis, M, Chang, G, Horton, N.C, Kercher, M.A, Pace, H.C, Lu, P. | | Deposit date: | 1996-01-03 | | Release date: | 1996-02-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.8 Å) | | Cite: | Crystal structure of the lactose operon repressor and its complexes with DNA and inducer.
Science, 271, 1996
|
|
4KSD
 
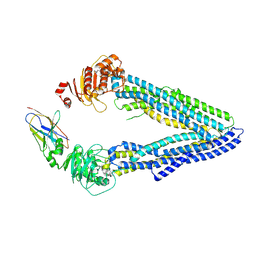 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A, R2 protein | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (4.1001 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KSC
 
 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4KSB
 
 | | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain | | Descriptor: | Multidrug resistance protein 1A | | Authors: | Ward, A, Szewczyk, P, Grimard, V, Lee, C.-W, Martinez, L, Doshi, R, Caya, A, Villaluz, M, Pardon, E, Cregger, C, Swartz, D.J, Falson, P, Urbatsch, I, Govaerts, C, Steyaert, J, Chang, G. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8001 Å) | | Cite: | Structures of P-glycoprotein reveal its conformational flexibility and an epitope on the nucleotide-binding domain.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2GPW
 
 | | Crystal Structure of the Biotin Carboxylase Subunit, F363A Mutant, of Acetyl-CoA Carboxylase from Escherichia coli. | | Descriptor: | Biotin carboxylase | | Authors: | Shen, Y, Chou, C.Y, Chang, G.G, Tong, L. | | Deposit date: | 2006-04-18 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Is dimerization required for the catalytic activity of bacterial biotin carboxylase?
Mol.Cell, 22, 2006
|
|
