4EZB
 
 | | CRYSTAL STRUCTURE OF the Conserved hypothetical protein from Sinorhizobium meliloti 1021 | | Descriptor: | uncharacterized conserved protein | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-05-02 | | Release date: | 2012-05-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF the Conserved hypothetical protein from Sinorhizobium meliloti 1021
To be Published
|
|
5IG3
 
 | | Crystal structure of the human CaMKII-alpha hub | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II subunit alpha | | Authors: | McSpadden, E, Cao, Y.M, Bhattacharyya, M, Gee, C.L, Barros, T, Kuriyan, J. | | Deposit date: | 2016-02-26 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular mechanism of activation-triggered subunit exchange in Ca(2+)/calmodulin-dependent protein kinase II.
Elife, 5, 2016
|
|
2VH0
 
 | | Structure and property based design of factor Xa inhibitors:biaryl pyrrolidin-2-ones incorporating basic heterocyclic motifs | | Descriptor: | 2-(5-chlorothiophen-2-yl)-N-[(3S)-1-(4-{2-[(dimethylamino)methyl]-1H-imidazol-1-yl}-2-fluorophenyl)-2-oxopyrrolidin-3-yl]ethanesulfonamide, ACTIVATED FACTOR XA HEAVY CHAIN, ACTIVATED FACTOR XA LIGHT CHAIN, ... | | Authors: | Young, R.J, Borthwick, A.D, Brown, D, Burns-Kurtis, C.L, Campbell, M, Chan, C, Charbaut, M, Convery, M.A, Diallo, H, Hortense, E, Irving, W.R, Kelly, H.A, King, N.P, Kleanthous, S, Mason, A.M, Pateman, A.J, Patikis, A, Pinto, I.L, Pollard, D.R, Senger, S, Shah, G.P, Toomey, J.R, Watson, N.S, Weston, H.E, Zhou, P. | | Deposit date: | 2007-11-16 | | Release date: | 2008-11-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and property based design of factor Xa inhibitors: biaryl pyrrolidin-2-ones incorporating basic heterocyclic motifs.
Bioorg. Med. Chem. Lett., 18, 2008
|
|
3I3G
 
 | | Crystal Structure of Trypanosoma brucei N-acetyltransferase (Tb11.01.2886) at 1.86A | | Descriptor: | N-acetyltransferase | | Authors: | Qiu, W, Wernimont, A.K, Marino, K, Zhang, A.Z, Ma, D, Lin, Y.H, Mackenzie, F, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, J Ferguson, M.A, Hui, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure Trypanosoma brucei N-acetyltransferase (Tb11.01.2886) at 1.86A
To be Published
|
|
1BG0
 
 | | TRANSITION STATE STRUCTURE OF ARGININE KINASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE KINASE, D-ARGININE, ... | | Authors: | Zhou, G, Somasundaram, T, Blanc, E, Parthasarathy, G, Ellington, W.R, Chapman, M.S. | | Deposit date: | 1998-06-03 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Transition state structure of arginine kinase: implications for catalysis of bimolecular reactions.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
3CLV
 
 | | Crystal Structure of Rab5a from plasmodium falciparum, PFB0500c | | Descriptor: | CHLORIDE ION, GUANOSINE-5'-DIPHOSPHATE, Rab5 protein, ... | | Authors: | Chattopadhyay, D, Wernimont, A.K, Langsley, G, Lew, J, Kozieradzki, I, Cossar, D, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Sukumar, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-03-20 | | Release date: | 2008-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Crystal Structure of Rab5a from plasmodium falciparum, PFB0500c
To be Published
|
|
7MS7
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (5-((4-(4-chlorophenyl)piperidin-1-yl)sulfonyl)picolinoyl)glycine | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, N-{5-[4-(4-chlorophenyl)piperidine-1-sulfonyl]pyridine-2-carbonyl}glycine, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M. | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7MS5
 
 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with 4-(4-(4-(3,4-difluoro-phenyl)-piperidin-1-ylsulfonyl)-phenyl)-4-oxo-butanoic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-[4-(3,4-difluorophenyl)piperidine-1-sulfonyl]phenyl}-4-oxobutanoic acid, CALCIUM ION, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
7MS6
 
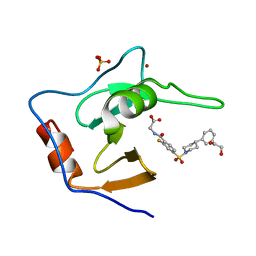 | | Structure of USP5 zinc-finger ubiquitin binding domain co-crystallized with (2-fluoro-4-((4-phenylpiperidin-1-yl)sulfonyl)benzoyl)glycine | | Descriptor: | 1,2-ETHANEDIOL, N-[2-fluoro-4-(4-phenylpiperidine-1-sulfonyl)benzoyl]glycine, SULFATE ION, ... | | Authors: | Mann, M.K, Zepeda-Velazquez, C.A, Alvarez, H.G, Dong, A, Kiyota, T, Aman, A, Arrowsmith, C.H, Al-Awar, R, Harding, R.J, Schapira, M. | | Deposit date: | 2021-05-10 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Activity Relationship of USP5 Inhibitors.
J.Med.Chem., 64, 2021
|
|
1AP4
 
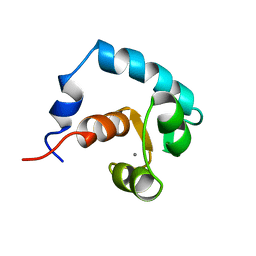 | | REGULATORY DOMAIN OF HUMAN CARDIAC TROPONIN C IN THE CALCIUM-SATURATED STATE, NMR, 40 STRUCTURES | | Descriptor: | CALCIUM ION, CARDIAC N-TROPONIN C | | Authors: | Li, M.X, Spyracopoulos, L, Sia, S.K, Gagne, S.M, Chandra, M, Solaro, R.J, Sykes, B.D. | | Deposit date: | 1997-07-24 | | Release date: | 1998-07-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Calcium-induced structural transition in the regulatory domain of human cardiac troponin C.
Biochemistry, 36, 1997
|
|
4P52
 
 | | Crystal structure of homoserine kinase from Cytophaga hutchinsonii ATCC 33406, NYSGRC Target 032717. | | Descriptor: | Homoserine kinase | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-03-13 | | Release date: | 2014-04-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of homoserine kinase from Cytophaga hutchinsonii ATCC 33406, NYSGRC Target 032717.
to be published
|
|
3J4B
 
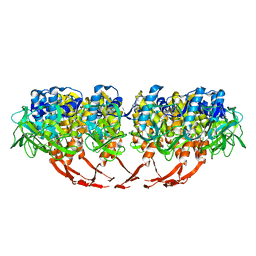 | | Structure of T7 gatekeeper protein (gp11) | | Descriptor: | Tail tubular protein A | | Authors: | Cuervo, A, Pulido-Cid, M, Chagoyen, M, Arranz, R, Gonzalez-Garcia, V.A, Garcia-Doval, C, Caston, J.R, Valpuesta, J.M, van Raaij, M.J, Martin-Benito, J, Carrascosa, J.L. | | Deposit date: | 2013-07-09 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural characterization of the bacteriophage t7 tail machinery.
J.Biol.Chem., 288, 2013
|
|
7LMT
 
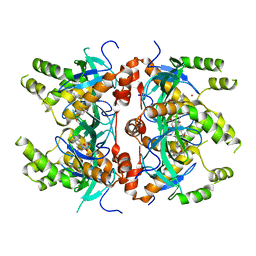 | | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound MRT10241866a | | Descriptor: | Histone-lysine N-methyltransferase NSD2, UNKNOWN ATOM OR ION, ~{N}-cyclopropyl-3-oxidanylidene-~{N}-(thiophen-2-ylmethyl)-4~{H}-1,4-benzoxazine-7-carboxamide | | Authors: | Lei, M, Freitas, R.F, Dong, A, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-02-05 | | Release date: | 2021-03-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Histone-lysine N-methyltransferase NSD2-PWWP1 with compound MRT10241866a
To Be Published
|
|
5IT6
 
 | | Galectin-related protein: an integral member of the network of chicken galectins | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Galectin-related protein, ... | | Authors: | Flores-Ibarra, A, Garcia-Caballero, G, Michalak, M, Bovin, N.V, Andre, S, Manning, J.C, Vertesy, S, Ruiz, F.M, Kaltner, H, Kopitz, J, Romero, A, Gabius, H.-J. | | Deposit date: | 2016-03-16 | | Release date: | 2016-07-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Galectin-related protein: An integral member of the network of chicken galectins 1. From strong sequence conservation of the gene confined to vertebrates to biochemical characteristics of the chicken protein and its crystal structure.
Biochim.Biophys.Acta, 1860, 2016
|
|
5IQM
 
 | | Crystal structure of the E. coli type 1 pilus subunit FimG (engineered variant with substitution Q134E; N-terminal FimG residues 1-12 truncated) in complex with the donor strand peptide DsF_T4R-T6R-D13N | | Descriptor: | COBALT (II) ION, Protein FimF, Protein FimG | | Authors: | Giese, C, Eras, J, Kern, A, Scharer, M.A, Capitani, G, Glockshuber, R. | | Deposit date: | 2016-03-11 | | Release date: | 2016-07-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Accelerating the Association of the Most Stable Protein-Ligand Complex by More than Two Orders of Magnitude.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
2K1K
 
 | |
6LFP
 
 | |
6EGX
 
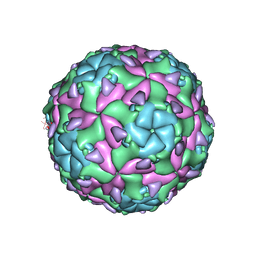 | | Sacbrood virus of honeybee - expansion state I | | Descriptor: | minor capsid protein MiCP, structural protein VP1, structural protein VP2, ... | | Authors: | Plevka, P, Prochazkova, M. | | Deposit date: | 2017-09-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Virion structure and genome delivery mechanism of sacbrood honeybee virus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4HWU
 
 | | Crystal structure of the Ig-C2 type 1 domain from mouse Fibroblast growth factor receptor 2 (FGFR2) [NYSGRC-005912] | | Descriptor: | Fibroblast growth factor receptor 2 | | Authors: | Kumar, P.R, Ahmed, M, Banu, R, Bhosle, R, Calarese, D, Celikigil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S, Glenn, A.S, Hillerich, B, Khafizov, K, Love, J, Patel, H, Rubinstein, R, Seidel, R, Stead, M, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2012-11-08 | | Release date: | 2012-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Crystal structure of the Ig-C2 type 1 domain from mouse FGFR2 [NYSGRC-005912]
to be published
|
|
6I6C
 
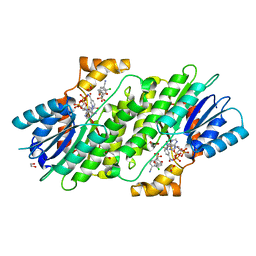 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 2 | | Descriptor: | (1~{R},2~{S},4~{S})-~{N}-(3-chloranyl-4-cyano-phenyl)sulfonylbicyclo[2.2.1]heptane-2-carboxamide, 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
6EO1
 
 | | The electron crystallography structure of the cAMP-bound potassium channel MloK1 (PCO-refined) | | Descriptor: | Cyclic nucleotide-gated potassium channel mll3241, POTASSIUM ION | | Authors: | Kowal, J, Biyani, N, Chami, M, Scherer, S, Rzepiela, A, Baumgartner, P, Upadhyay, V, Nimigean, C, Stahlberg, H. | | Deposit date: | 2017-10-08 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (4.5 Å) | | Cite: | High-Resolution Cryoelectron Microscopy Structure of the Cyclic Nucleotide-Modulated Potassium Channel MloK1 in a Lipid Bilayer.
Structure, 26, 2018
|
|
6I6P
 
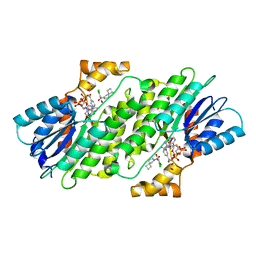 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 3 | | Descriptor: | 1,2-ETHANEDIOL, 6-azaspiro[3.4]octan-6-yl-[2,4-bis(chloranyl)-6-oxidanyl-phenyl]methanone, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
3H80
 
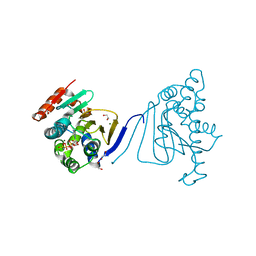 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LmjF33.0312:M1-K213 | | Descriptor: | 1,2-ETHANEDIOL, Heat shock protein 83-1, MAGNESIUM ION, ... | | Authors: | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, Mackenzie, F, Fairlamb, A, Kozieradzki, I, Cossar, D, Zhao, Y, Schapira, M, Bochkarev, A, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-28 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LmjF33.0312:M1-K213
To be Published
|
|
5J9A
 
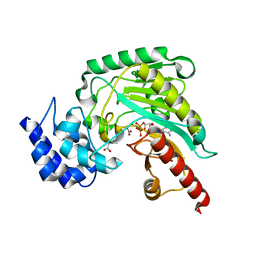 | | Ambient temperature transition state structure of arginine kinase - crystal 11/Form II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ARGININE, Arginine kinase, ... | | Authors: | Godsey, M, Davulcu, O, Nix, J, Skalicky, J.J, Bruschweiler, R, Chapman, M.S. | | Deposit date: | 2016-04-08 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | The Sampling of Conformational Dynamics in Ambient-Temperature Crystal Structures of Arginine Kinase.
Structure, 24, 2016
|
|
6EH1
 
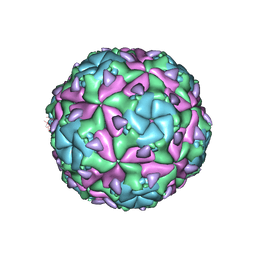 | | Sacbrood virus of honeybee - expansion state II | | Descriptor: | minor capsid protein MiCP, structural protein VP1, structural protein VP2, ... | | Authors: | Plevka, P, Prochazkova, M. | | Deposit date: | 2017-09-12 | | Release date: | 2018-07-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.25 Å) | | Cite: | Virion structure and genome delivery mechanism of sacbrood honeybee virus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
