1QZ8
 
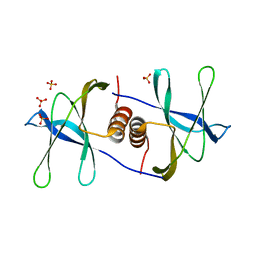 | | Crystal structure of SARS coronavirus NSP9 | | Descriptor: | SULFATE ION, polyprotein 1ab | | Authors: | Egloff, M.P, Ferron, F, Campanacci, V, Longhi, S, Rancurel, C, Dutartre, H, Snijder, E.J, Gorbalenya, A.E, Cambillau, C, Canard, B. | | Deposit date: | 2003-09-16 | | Release date: | 2004-02-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The severe acute respiratory syndrome-coronavirus replicative protein nsp9 is a single-stranded RNA-binding subunit unique in the RNA virus world.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6YU8
 
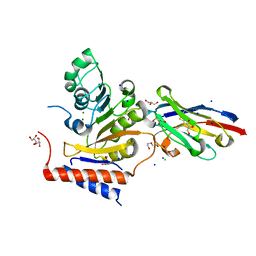 | | RNA Methyltransferase of Sudan Ebola Virus | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Ferron, F, Valle, C, Zamboni, V, Canard, B, Decroly, E. | | Deposit date: | 2020-04-25 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | First insights into the structural features of Ebola virus methyltransferase activities.
Nucleic Acids Res., 49, 2021
|
|
8QCH
 
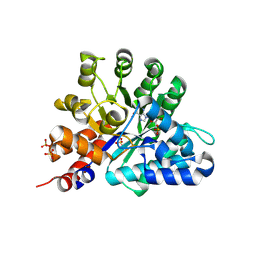 | | Human Adenosine deaminase-like protein in complex with compound AT8001 | | Descriptor: | Adenosine deaminase-like protein, DI(HYDROXYETHYL)ETHER, SODIUM ION, ... | | Authors: | Zimberger, C, canard, B, Ferron, F. | | Deposit date: | 2023-08-26 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.442 Å) | | Cite: | The activation cascade of the broad-spectrum antiviral bemnifosbuvir characterized at atomic resolution.
Plos Biol., 22, 2024
|
|
1R6A
 
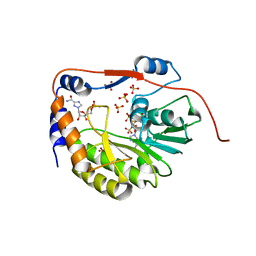 | | Structure of the dengue virus 2'O methyltransferase in complex with s-adenosyl homocysteine and ribavirin 5' triphosphate | | Descriptor: | Genome polyprotein, RIBAVIRIN MONOPHOSPHATE, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Benarroch, D, Egloff, M.P, Mulard, L, Romette, J.L, Canard, B. | | Deposit date: | 2003-10-15 | | Release date: | 2004-09-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A structural basis for the inhibition of the NS5 dengue virus mRNA 2'-O-methyltransferase domain by ribavirin 5'-triphosphate.
J.Biol.Chem., 279, 2004
|
|
8CB3
 
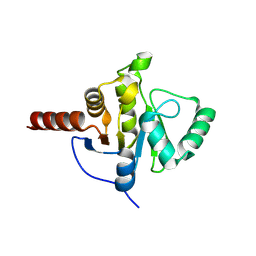 | | SARS-CoV Macro domain complexed with 3-(N-morpholino)propanesulfonic acid | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, GLYCEROL, Papain-like protease nsp3 | | Authors: | Morin, B, Ferron, F, Coutard, B, Canard, B, Marseilles Structural Genomics Program @ AFMB (MSGP) | | Deposit date: | 2023-01-25 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | SARS-CoV Macro domain complexed with 3-(N-morpholino)propanesulfonic acid
To Be Published
|
|
6G13
 
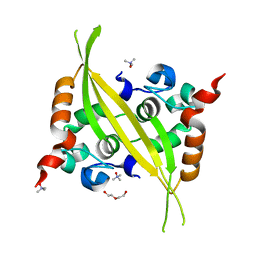 | | C-terminal domain of MERS-CoV nucleocapsid | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Nucleoprotein, ... | | Authors: | Nguyen, T.H.V, Ferron, F.P, Lichiere, J, Canard, B, Papageorgiou, N, Coutard, B. | | Deposit date: | 2018-03-20 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure and oligomerization state of the C-terminal region of the Middle East respiratory syndrome coronavirus nucleoprotein.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7ED5
 
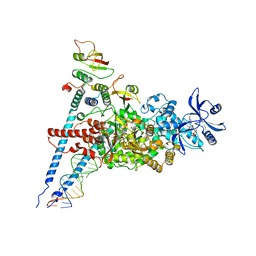 | | A dual mechanism of action of AT-527 against SARS-CoV-2 polymerase | | Descriptor: | MAGNESIUM ION, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Shannon, A, Fattorini, V, Sama, B, Selisko, B, Feracci, M, Falcou, C, Gauffre, P, El Kazzi, P, Delpal, A, Decroly, E, Alvarez, K, Eydoux, C, Guillemot, J.-C, Moussa, A, Good, S, Colla, P, Lin, K, Sommadossi, J.-P, Zhu, Y.X, Yan, X.D, Shi, H, Ferron, F, Canard, B. | | Deposit date: | 2021-03-15 | | Release date: | 2022-02-16 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | A dual mechanism of action of AT-527 against SARS-CoV-2 polymerase.
Nat Commun, 13, 2022
|
|
1OKS
 
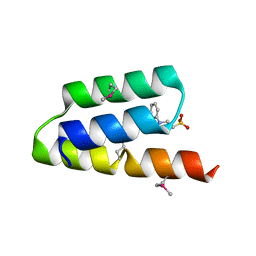 | | Crystal structure of the measles virus phosphoprotein XD domain | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, RNA POLYMERASE ALPHA SUBUNIT | | Authors: | Johansson, K, Bourhis, J.-M, Campanacci, V, Cambillau, C, Canard, B, Longhi, S. | | Deposit date: | 2003-07-29 | | Release date: | 2003-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Measles Virus Phosphoprotein Domain Responsible for the Induced Folding of the C-Terminal Domain of the Nucleoprotein
J.Biol.Chem., 278, 2003
|
|
1L9K
 
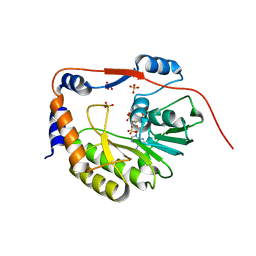 | | dengue methyltransferase | | Descriptor: | RNA-DIRECTED RNA POLYMERASE, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Egloff, M.P, Benarroch, D, Selisko, B, Romette, J.L, Canard, B. | | Deposit date: | 2002-03-25 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | An RNA cap (nucleoside-2'-O-) methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization
Embo J., 21, 2002
|
|
8C0G
 
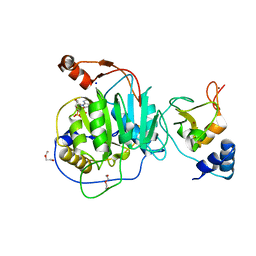 | | SARS-CoV nsp16-nsp10 complexed with N7-GTP | | Descriptor: | DI(HYDROXYETHYL)ETHER, Non-structural protein 7, SODIUM ION, ... | | Authors: | Ferron, F, Debarnot, C, Coutard, B, Canard, B, Marseilles Structural Genomics Program @ AFMB (MSGP) | | Deposit date: | 2022-12-16 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Observation of N7-GTP in nsp16-nsp10 Sars-CoV
To Be Published
|
|
8BCR
 
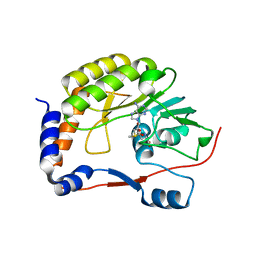 | | DENV3 Methyltransferase in complexed with AT-9010 and SAH | | Descriptor: | Genome polyprotein, PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Gauffre, P, Fattorini, V, Canard, B, Ferron, F. | | Deposit date: | 2022-10-17 | | Release date: | 2023-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | AT-752 targets multiple sites and activities on the Dengue virus replication enzyme NS5.
Antiviral Res., 212, 2023
|
|
8PWK
 
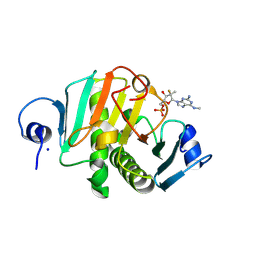 | | human HINT1 in complex with compound AT8003 | | Descriptor: | Histidine triad nucleotide-binding protein 1, SODIUM ION, [(2~{R},3~{R},4~{R},5~{R})-5-[2-azanyl-6-(methylamino)purin-9-yl]-4-fluoranyl-4-methyl-3-oxidanyl-oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Zimberger, C, Canard, B, Ferron, F. | | Deposit date: | 2023-07-20 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | The activation cascade of the broad-spectrum antiviral bemnifosbuvir characterized at atomic resolution.
Plos Biol., 22, 2024
|
|
8PTS
 
 | | human GUK1 in complex with compound AT8001 | | Descriptor: | GLYCEROL, Guanylate kinase, [(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-4-fluoranyl-4-methyl-3-oxidanyl-oxolan-2-yl]methyl dihydrogen phosphate | | Authors: | Zimberger, C, Canard, B, Ferron, F. | | Deposit date: | 2023-07-14 | | Release date: | 2024-07-24 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The activation cascade of the broad-spectrum antiviral bemnifosbuvir characterized at atomic resolution.
Plos Biol., 22, 2024
|
|
8PIE
 
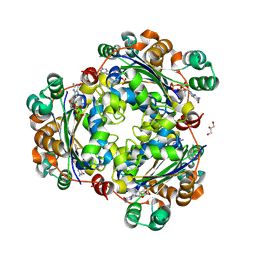 | | Crystal structure of the human nucleoside diphosphate kinase B domain in complex with the product AT-8500 formed by catalysis of compound AT-9010 | | Descriptor: | GLYCEROL, Nucleoside diphosphate kinase B, [(2~{R},3~{R},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-4-fluoranyl-4-methyl-3-oxidanyl-oxolan-2-yl]methyl phosphono hydrogen phosphate | | Authors: | Feracci, M, Chazot, A, Ferron, F, Alvarez, K, Canard, B. | | Deposit date: | 2023-06-21 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The activation cascade of the broad-spectrum antiviral bemnifosbuvir characterized at atomic resolution.
Plos Biol., 22, 2024
|
|
2CKW
 
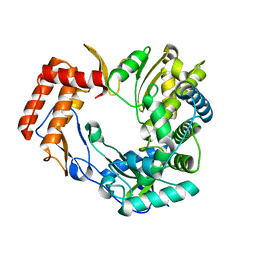 | | The 2.3 A resolution structure of the Sapporo virus RNA dependant RNA polymerase. | | Descriptor: | RNA-DIRECTED RNA POLYMERASE | | Authors: | Fullerton, S.W.B, Tucker, P.A, Rohayem, J, Coutard, B, Gebhardt, J, Gorbalenya, A, Canard, B. | | Deposit date: | 2006-04-24 | | Release date: | 2006-11-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Characterization of Sapovirus RNA-Dependent RNA Polymerase.
J.Virol., 81, 2007
|
|
5T2T
 
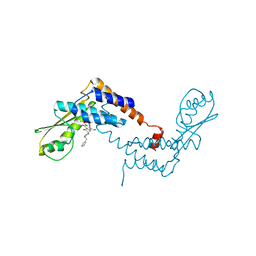 | | Crystal structure of Lymphocytic choriomeningitis mammarenavirus endonuclease bound to compound L742001 | | Descriptor: | (2Z)-4-[1-benzyl-4-(4-chlorobenzyl)piperidin-4-yl]-2-hydroxy-4-oxobut-2-enoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Saez-Ayala, M, Yekwa, E.L, Canard, B, Alvarez, K, Ferron, F. | | Deposit date: | 2016-08-24 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.967 Å) | | Cite: | Crystal structures ofLymphocytic choriomeningitis virusendonuclease domain complexed with diketo-acid ligands.
IUCrJ, 5, 2018
|
|
7OA4
 
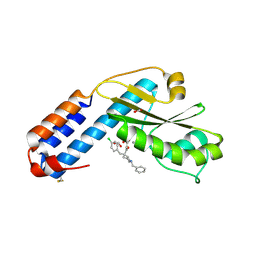 | | Crystal structure of the N-terminal endonuclease domain of La Crosse virus L-protein bound to compound L-742,001 | | Descriptor: | (2Z)-4-[1-benzyl-4-(4-chlorobenzyl)piperidin-4-yl]-2-hydroxy-4-oxobut-2-enoic acid, FORMIC ACID, MANGANESE (II) ION, ... | | Authors: | Feracci, M, Hernandez, S, Vincentelli, R, Ferron, F, Reguera, J, Canard, B, Alvarez, K. | | Deposit date: | 2021-04-19 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Biophysical and structural study of La Crosse virus endonuclease inhibition for the development of new antiviral options.
Iucrj, 11, 2024
|
|
5FVA
 
 | | Toscana Virus Nucleocapsid Protein | | Descriptor: | NUCLEOPROTEIN, SODIUM ION | | Authors: | Baklouti, A, Sevajol, M, Goulet, A, Lichiere, J, Ferron, F, Canard, B, Charrel, R.N, Coutard, B, Papageorgiou, N. | | Deposit date: | 2016-02-03 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Toscana virus nucleoprotein oligomer organization observed in solution.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
7Z2J
 
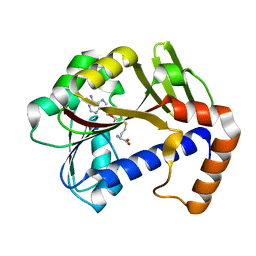 | | White Bream virus N7-Methyltransferase | | Descriptor: | Non-structural protein 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Shannon, A, Gauffre, P, Canard, B, Ferron, F. | | Deposit date: | 2022-02-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.657 Å) | | Cite: | A second type of N7-guanine RNA cap methyltransferase in an unusual locus of a large RNA virus genome.
Nucleic Acids Res., 50, 2022
|
|
7Z05
 
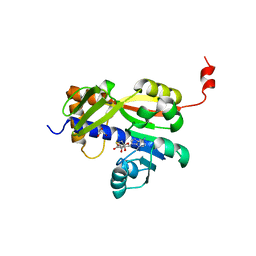 | | White Bream virus N7-Methyltransferase | | Descriptor: | DI(HYDROXYETHYL)ETHER, Non-structural protein 1, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Shannon, A, Gauffre, P, Canard, B, Ferron, F. | | Deposit date: | 2022-02-22 | | Release date: | 2022-09-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | A second type of N7-guanine RNA cap methyltransferase in an unusual locus of a large RNA virus genome.
Nucleic Acids Res., 50, 2022
|
|
1MN9
 
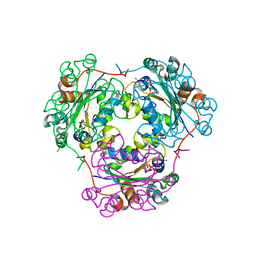 | | NDP kinase mutant (H122G) complex with RTP | | Descriptor: | MAGNESIUM ION, NDP kinase, RIBAVIRIN TRIPHOSPHATE | | Authors: | Gallois-montbrun, S, Chen, Y, Dutartre, H, Morera, S, Guerreiro, C, Mulard, L, Schneider, B, Janin, J, Canard, B, Veron, M, Deville-bonne, D. | | Deposit date: | 2002-09-05 | | Release date: | 2003-03-18 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Analysis of the Activation of Ribavirin Analogs by NDP Kinase: Comparison with Other Ribavirin Targets
MOL.PHARMACOL., 63, 2003
|
|
1F6T
 
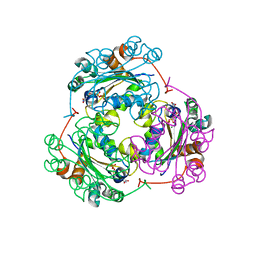 | | STRUCTURE OF THE NUCLEOSIDE DIPHOSPHATE KINASE/ALPHA-BORANO(RP)-TDP.MG COMPLEX | | Descriptor: | 2*-DEOXY-THYMIDINE-5*-ALPHA BORANO DIPHOSPHATE (ISOMER RP), MAGNESIUM ION, PROTEIN (NUCLEOSIDE DIPHOSPHATE KINASE) | | Authors: | Guerreiro, C, Boretto, J, Janin, J, Veron, M, Canard, B, Schneider, B, Sarfati, S, Deville-Bonne, D. | | Deposit date: | 2000-06-23 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for activation of alpha-boranophosphate nucleotide analogues targeting drug-resistant reverse transcriptase.
EMBO J., 19, 2000
|
|
1F3F
 
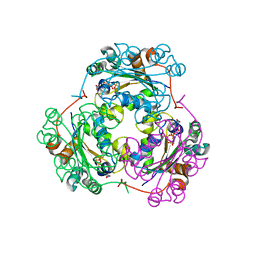 | | STRUCTURE OF THE H122G NUCLEOSIDE DIPHOSPHATE KINASE / D4T-TRIPHOSPHATE.MG COMPLEX | | Descriptor: | 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-DIPHOSPHATE, 2',3'-DEHYDRO-2',3'-DEOXY-THYMIDINE 5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Meyer, P, Schneider, B, Sarfati, S, Deville-Bonne, D, Guerreiro, C, Boretto, J, Janin, J, Veron, M, Canard, B. | | Deposit date: | 2000-06-02 | | Release date: | 2000-09-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for activation of alpha-boranophosphate nucleotide analogues targeting drug-resistant reverse transcriptase.
EMBO J., 19, 2000
|
|
2XYQ
 
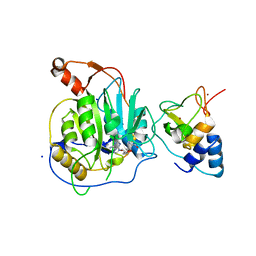 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-18 | | Release date: | 2011-10-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
2XYV
 
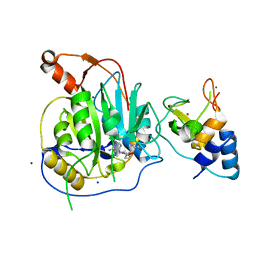 | | Crystal structure of the nsp16 nsp10 SARS coronavirus complex | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NON-STRUCTURAL PROTEIN 10, ... | | Authors: | Decroly, E, Debarnot, C, Ferron, F, Bouvet, M, Coutard, B, Imbert, I, Gluais, L, Papageorgiou, N, Ortiz-Lombardia, M, Lescar, J, Canard, B. | | Deposit date: | 2010-11-19 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure and Functional Analysis of the Sars-Coronavirus RNA CAP 2'-O-Methyltransferase Nsp10/Nsp16 Complex.
Plos Pathog., 7, 2011
|
|
