4LUP
 
 | | Crystal structure of the complex formed by region of E. coli sigmaE bound to its -10 element non template strand | | Descriptor: | 1,2-ETHANEDIOL, RNA polymerase sigma factor, region 2 of sigmaE of E. coli | | Authors: | Campagne, S, Marsh, M.E, Vorholt, J.A.V, Allain, F.H.-T, Capitani, G. | | Deposit date: | 2013-07-25 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis for -10 promoter element melting by environmentally induced sigma factors.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5OR5
 
 | |
5OR0
 
 | |
7AEP
 
 | | Solution structure of U1-A RRM2 (190-282) | | Descriptor: | U1 small nuclear ribonucleoprotein A | | Authors: | Campagne, S, Allain, F.H. | | Deposit date: | 2020-09-18 | | Release date: | 2021-02-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | An in vitro reconstituted U1 snRNP allows the study of the disordered regions of the particle and the interactions with proteins and ligands.
Nucleic Acids Res., 49, 2021
|
|
6SNJ
 
 | |
7ZAP
 
 | |
7Q33
 
 | |
3K16
 
 | | Crystal Structure of BRCA1 BRCT D1840T in complex with a minimal recognition tetrapeptide with a free carboxy C-terminus | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-25 | | Release date: | 2010-03-02 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
3K0H
 
 | | The crystal structure of BRCA1 BRCT in complex with a minimal recognition tetrapeptide with an amidated C-terminus | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
3K15
 
 | | Crystal Structure of BRCA1 BRCT D1840T in complex with a minimal recognition tetrapeptide with an amidated C-terminus | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-25 | | Release date: | 2010-03-02 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
3K05
 
 | |
3K0K
 
 | | Crystal Structure of BRCA1 BRCT in complex with a minimal recognition tetrapeptide with a free carboxy C-terminus. | | Descriptor: | Breast cancer type 1 susceptibility protein, CHLORIDE ION, NICKEL (II) ION, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N. | | Deposit date: | 2009-09-24 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Comparison of the Structures and Peptide Binding Specificities of the BRCT Domains of MDC1 and BRCA1
Structure, 18, 2010
|
|
6HMI
 
 | |
2JOS
 
 | |
2LFW
 
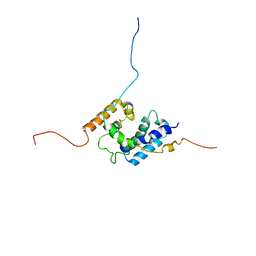 | | NMR structure of the PhyRSL-NepR complex from Sphingomonas sp. Fr1 | | Descriptor: | NepR anti sigma factor, PhyR sigma-like domain | | Authors: | Campagne, S, Damberger, F.F, Vorholt, J.A, Allain, F.H.-T. | | Deposit date: | 2011-07-18 | | Release date: | 2012-04-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sigma factor mimicry in the general stress response of Alphaproteobacteria.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2MAO
 
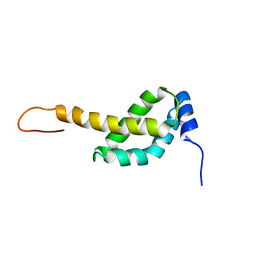 | |
2MAP
 
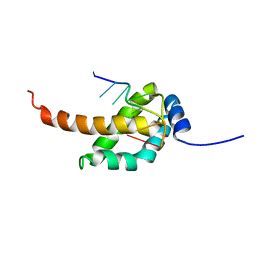 | |
6HMO
 
 | |
2L1G
 
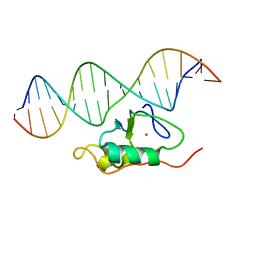 | | RDC refined solution structure of the THAP zinc finger of THAP1 in complex with its 16bp RRM1 DNA target | | Descriptor: | DNA (5'-D(*GP*CP*TP*TP*GP*TP*GP*TP*GP*GP*GP*CP*AP*GP*CP*G)-3'), DNA (5'-D(P*CP*GP*CP*TP*GP*CP*CP*CP*AP*CP*AP*CP*AP*AP*GP*C)-3'), THAP domain-containing protein 1, ... | | Authors: | Campagne, S, Gervais, V, Saurel, O, Milon, A. | | Deposit date: | 2010-07-28 | | Release date: | 2010-09-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | RDC refined solution structure of the THAP zinc finger of THAP1 in complex with its 16bp RRM1 DNA target
To be published
|
|
5IEJ
 
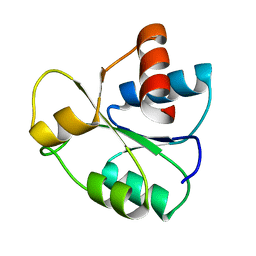 | |
5IEB
 
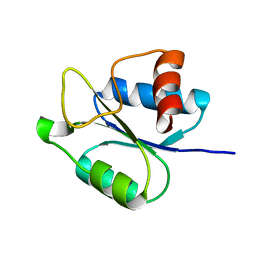 | |
2KO0
 
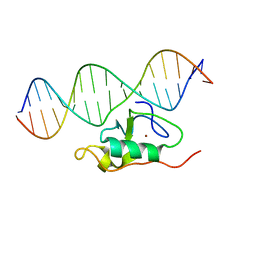 | | Solution structure of the THAP zinc finger of THAP1 in complex with its DNA target | | Descriptor: | RRM1, THAP domain-containing protein 1, ZINC ION | | Authors: | Campagne, S, Gervais, V, Saurel, O, Milon, A. | | Deposit date: | 2009-09-08 | | Release date: | 2010-01-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural determinants of specific DNA-recognition by the THAP zinc finger
Nucleic Acids Res., 2010
|
|
4ORH
 
 | | Crystal structure of RNF8 bound to the UBC13/MMS2 heterodimer | | Descriptor: | E3 ubiquitin-protein ligase RNF8, Ubiquitin-conjugating enzyme E2 N, Ubiquitin-conjugating enzyme E2 variant 2, ... | | Authors: | Campbell, S.J, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2014-02-11 | | Release date: | 2014-02-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.802 Å) | | Cite: | Molecular insights into the function of RING finger (RNF)-containing proteins hRNF8 and hRNF168 in Ubc13/Mms2-dependent ubiquitylation.
J.Biol.Chem., 287, 2012
|
|
2NB9
 
 | | Solution structure of ZitP zinc finger | | Descriptor: | Uncharacterized protein, ZINC ION | | Authors: | Campagne, S, Berge, M, Viollier, P.H, Allain, F.H.-T. | | Deposit date: | 2016-02-01 | | Release date: | 2016-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Modularity and determinants of a (bi-)polarization control system from free-living and obligate intracellular bacteria.
Elife, 5, 2016
|
|
4EQF
 
 | | Trip8b-1a#206-567 interacting with the carboxy-terminal seven residues of HCN2 | | Descriptor: | PEX5-related protein, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Bankston, J.R, Camp, S.S, Dimaio, F, Lewis, A.S, Chetkovich, D.M, Zagotta, W.N. | | Deposit date: | 2012-04-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure and stoichiometry of an accessory subunit TRIP8b interaction with hyperpolarization-activated cyclic nucleotide-gated channels.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
