6LDX
 
 | |
6LDY
 
 | |
6LDW
 
 | |
6LDV
 
 | |
6K2G
 
 | |
7CUE
 
 | | Crystal structure of HID2 bound to human Hemoglobin | | Descriptor: | Amino acid ABC transporter substrate-binding protein, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Caaveiro, J.M.M, Hoshino, M, Tsumoto, K. | | Deposit date: | 2020-08-22 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the recognition of human hemoglobin by the Shr protein from Streptococcus pyogenes
To Be Published
|
|
7CUD
 
 | |
4TSO
 
 | | Crystal structure of FraC with DHPC bound (crystal form I) | | Descriptor: | 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Fragaceatoxin C, PHOSPHATE ION, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
4TSP
 
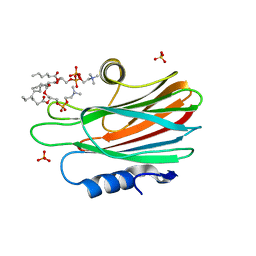 | | Crystal structure of FraC with DHPC bound (crystal form II) | | Descriptor: | 1,2-dihexanoyl-sn-glycero-3-phosphocholine, Fragaceatoxin C, PHOSPHATE ION, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
4TSL
 
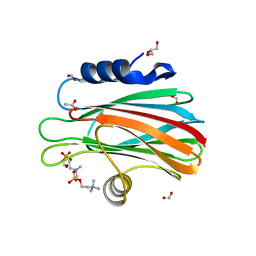 | | Crystal structure of FraC with POC bound (crystal form I) | | Descriptor: | ACETATE ION, FORMIC ACID, Fragaceatoxin C, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
4TSY
 
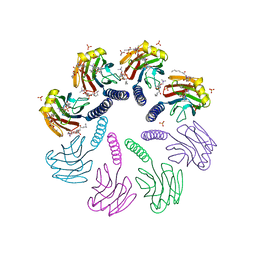 | | Crystal structure of FraC with lipids | | Descriptor: | 2-[[(E,2S,3R)-2-(hexanoylamino)-3-oxidanyl-dec-4-enoxy]-oxidanyl-phosphoryl]oxyethyl-trimethyl-azanium, Fragaceatoxin C, HEPTANE-1,2,3-TRIOL, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
7EV1
 
 | | Crystal structure of LI-Cadherin EC1-2 | | Descriptor: | ACETATE ION, CALCIUM ION, Cadherin-17, ... | | Authors: | Caaveiro, J.M.M, Yui, A, Tsumoto, K. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Mechanism of dimerization and structural features of human LI-cadherin.
J.Biol.Chem., 297, 2021
|
|
4TSN
 
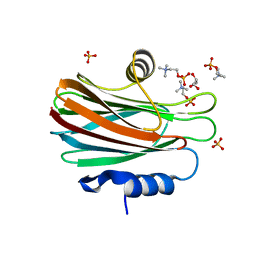 | | Crystal structure of FraC with POC bound (crystal form II) | | Descriptor: | ACETATE ION, Fragaceatoxin C, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
4TSQ
 
 | | Crystal structure of FraC with DHPC bound (crystal form III) | | Descriptor: | 1,2-dihexanoyl-sn-glycero-3-phosphocholine, CHLORIDE ION, Fragaceatoxin C, ... | | Authors: | Caaveiro, J.M.M, Tanaka, K, Tsumoto, K. | | Deposit date: | 2014-06-19 | | Release date: | 2015-03-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for self-assembly of a cytolytic pore lined by protein and lipid
Nat Commun, 6, 2015
|
|
7CYM
 
 | | Crystal structure of LI-Cadherin EC1-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Caaveiro, J.M.M, Yui, A, Tsumoto, K. | | Deposit date: | 2020-09-03 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Mechanism of dimerization and structural features of human LI-cadherin.
J.Biol.Chem., 297, 2021
|
|
7EKB
 
 | | Crystal structure of 4E10 modified with pyrene acetamide | | Descriptor: | ACETATE ION, Fab region of the heavy chain of broadly neutralizing antibody anti-HIV-1 4E10, Fab region of the light chain of the broadly neutralizing anti-HIV-1 antibody 4E10, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Focal accumulation of aromaticity at the CDRH3 loop mitigates 4E10 polyreactivity without altering its HIV neutralization profile.
Iscience, 24, 2021
|
|
7EKK
 
 | | Anti-HIV-1 broadly neutralizing antibody delta-loop 4E10 modified with pyrene acetamide | | Descriptor: | AMMONIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Caaveiro, J.M.M, Rujas, E, Nieva, J.L. | | Deposit date: | 2021-04-05 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Focal accumulation of aromaticity at the CDRH3 loop mitigates 4E10 polyreactivity without altering its HIV neutralization profile.
Iscience, 24, 2021
|
|
7W81
 
 | |
7WKI
 
 | |
7XAR
 
 | | Crystal structure of 3C-like protease from SARS-CoV-2 in complex with covalent inhibitor | | Descriptor: | 3C-like proteinase, 4-fluoranyl-~{N}-[(2~{S})-1-[2-(2-fluoranylethanoyl)-2-[[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]methyl]hydrazinyl]-4-methyl-1-oxidanylidene-pentan-2-yl]-1~{H}-indole-2-carboxamide, CHLORIDE ION, ... | | Authors: | Caaveiro, J.M.M, Ochi, J, Takahashi, D, Ueda, T, Ojida, A. | | Deposit date: | 2022-03-18 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Chlorofluoroacetamide-Based Covalent Inhibitors for Severe Acute Respiratory Syndrome Coronavirus 2 3CL Protease.
J.Med.Chem., 65, 2022
|
|
7XL0
 
 | | Crystal structure of Vobarilizumab at 1.70 Angstrom | | Descriptor: | GLYCEROL, Nanobody Vobarilizumab, SULFATE ION | | Authors: | Caaveiro, J.M.M, Mori, C, Kinoshita, S, Nakakido, M, Tsumoto, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis for thermal stability and affinity in a VHH: Contribution of the framework region and its influence in the conformation of the CDR3.
Protein Sci., 31, 2022
|
|
7XL1
 
 | | Crystal structure of chimeric 7D12-Vob nanobody at 1.65 Angstrom | | Descriptor: | Chimeric 7D12-Vob nanobody, MALONATE ION | | Authors: | Caaveiro, J.M.M, Kinoshita, S, Mori, C, Nakakido, M, Tsumoto, K. | | Deposit date: | 2022-04-20 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis for thermal stability and affinity in a VHH: Contribution of the framework region and its influence in the conformation of the CDR3.
Protein Sci., 31, 2022
|
|
7XLI
 
 | | Crystal structure of IsdB linker-NEAT2 bound to a nanobody (VHH) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Iron-regulated surface determinant protein B, ... | | Authors: | Caaveiro, J.M.M, Valenciano-Bellido, S, Tsumoto, K. | | Deposit date: | 2022-04-21 | | Release date: | 2023-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Targeting hemoglobin receptors IsdH and IsdB of Staphylococcus aureus with a single VHH antibody inhibits bacterial growth.
J.Biol.Chem., 299, 2023
|
|
7XLD
 
 | | Crystal structure of IsdH linker-NEAT3 bound to a nanobody (VHH) | | Descriptor: | Iron-regulated surface determinant protein H, MAGNESIUM ION, Nanobody VHH6, ... | | Authors: | Caaveiro, J.M.M, Valenciano-Bellido, S, Tsumoto, K. | | Deposit date: | 2022-04-21 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Targeting hemoglobin receptors IsdH and IsdB of Staphylococcus aureus with a single VHH antibody inhibits bacterial growth.
J.Biol.Chem., 299, 2023
|
|
5YD4
 
 | |
