7UXX
 
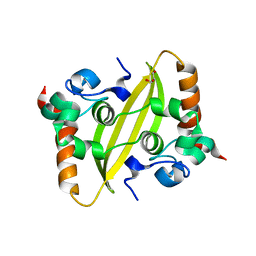 | | Crystal structure of SARS-CoV-2 nucleocapsid protein C-terminal domain | | Descriptor: | ACETATE ION, GLYCEROL, Nucleoprotein | | Authors: | Bezerra, E.H.S, Tonoli, C.C.C, Soprano, A.S, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Discovery and structural characterization of chicoric acid as a SARS-CoV-2 nucleocapsid protein ligand and RNA binding disruptor.
Sci Rep, 12, 2022
|
|
7UXZ
 
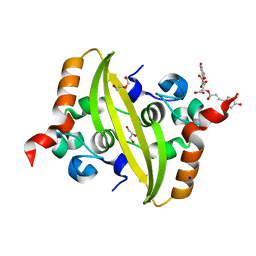 | | Crystal structure of SARS-CoV-2 nucleocapsid protein C-terminal domain complexed with Chicoric acid | | Descriptor: | (2R,3R)-2,3-bis{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}butanedioic acid, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Bezerra, E.H.S, Tonoli, C.C.C, Soprano, A.S, Franchini, K.G, Trivella, D.B.B, Benedetti, C.E. | | Deposit date: | 2022-05-06 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.733 Å) | | Cite: | Discovery and structural characterization of chicoric acid as a SARS-CoV-2 nucleocapsid protein ligand and RNA binding disruptor.
Sci Rep, 12, 2022
|
|
4COT
 
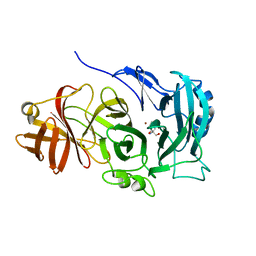 | | The importance of the Abn2 calcium cluster in the endo-1,5- arabinanase activity from Bacillus subtilis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, EXTRACELLULAR ENDO-ALPHA-(1->5)-L-ARABINANASE 2, NICKEL (II) ION | | Authors: | McVey, C.E, Ferreira, M.J, Correia, B, Lahiri, S, deSanctis, D, Carrondo, M.A, Lindley, P.F, de Sa-Nogueira, I, Soares, C.M, Bento, I. | | Deposit date: | 2014-01-31 | | Release date: | 2014-03-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Importance of the Abn2 Calcium Cluster in the Endo-1,5-Arabinanase Activity from Bacillus Subtilis.
J.Biol.Inorg.Chem., 19, 2014
|
|
7UW1
 
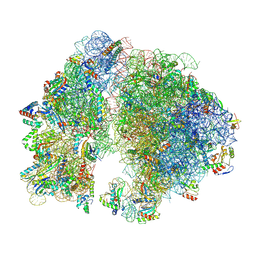 | | A. baumannii 70S ribosome-Streptothricin-D complex | | Descriptor: | 16s Ribosomal RNA, 23s ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2022-05-02 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Streptothricin F is a bactericidal antibiotic effective against highly drug-resistant gram-negative bacteria that interacts with the 30S subunit of the 70S ribosome.
Plos Biol., 21, 2023
|
|
4BHU
 
 | | Crystal structure of BslA - A bacterial hydrophobin | | Descriptor: | CHLORIDE ION, GLYCEROL, UNCHARACTERIZED PROTEIN YUAB | | Authors: | Rao, F.V, Hobley, L, Ostrowski, A, Bromley, K.M, Porter, M, Prescott, A.R, Swedlow, J.R, MacPhee, C.E, van Aalten, D.M.F, Stanley-Wall, N.R. | | Deposit date: | 2013-04-08 | | Release date: | 2013-08-14 | | Last modified: | 2013-08-28 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Bsla is a Self-Assembling Bacterial Hydrophobin that Coats the Bacillus Subtilis Biofilm.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4BLG
 
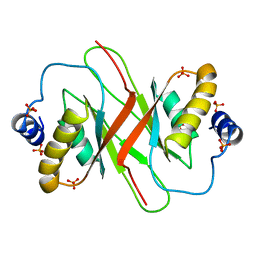 | | Crystal structure of MHV-68 Latency-associated nuclear antigen (LANA) C-terminal DNA binding domain | | Descriptor: | LATENCY-ASSOCIATED NUCLEAR ANTIGEN, PHOSPHATE ION | | Authors: | Correia, B, Cerqueira, S.A, Beauchemin, C, Pires De Miranda, M, Li, S, Ponnusamy, R, Rodrigues, L, Schneider, T.R, Carrondo, M.A, Kaye, K.M, Simas, J.P, McVey, C.E. | | Deposit date: | 2013-05-02 | | Release date: | 2013-10-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Gamma-2 Herpesvirus Lana DNA Binding Domain Identifies Charged Surface Residues which Impact Viral Latency
Plos Pathog., 9, 2013
|
|
4JS5
 
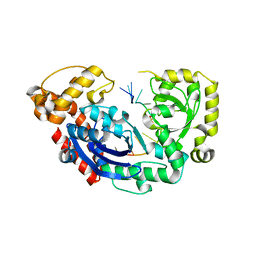 | |
4JS4
 
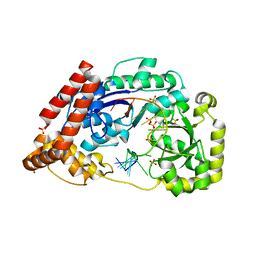 | |
4JRP
 
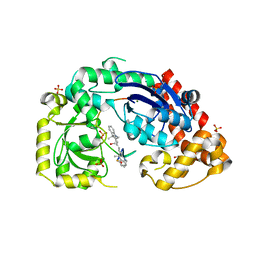 | | Structure of E. coli Exonuclease I in complex with a 5cy-dT13 oligonucleotide | | Descriptor: | 5cy-dT13, Exodeoxyribonuclease I, GLYCEROL, ... | | Authors: | Bell, C.E. | | Deposit date: | 2013-03-21 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of Escherichia coli exonuclease I in complex with single-stranded DNA provide insights into the mechanism of processive digestion.
Nucleic Acids Res., 41, 2013
|
|
4JRQ
 
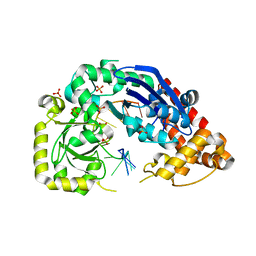 | |
2PXG
 
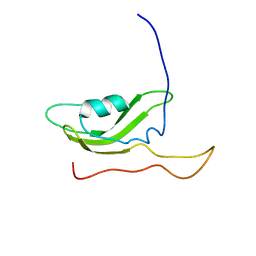 | | NMR Solution Structure of OmlA | | Descriptor: | Outer membrane protein | | Authors: | Vanini, M.M.T, Pertinhez, T.A, Sforca, M.L, Spisni, A, Benedetti, C.E. | | Deposit date: | 2007-05-14 | | Release date: | 2008-01-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the outer membrane lipoprotein OmlA from Xanthomonas axonopodis pv. citri reveals a protein fold implicated in protein-protein interaction.
Proteins, 71, 2008
|
|
2WTB
 
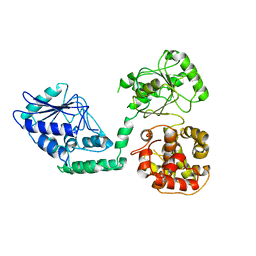 | | Arabidopsis thaliana multifuctional protein, MFP2 | | Descriptor: | FATTY ACID MULTIFUNCTIONAL PROTEIN (ATMFP2) | | Authors: | Arent, S, Pye, V.E, Christensen, C.E, Norgaard, A, Henriksen, A. | | Deposit date: | 2009-09-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Multi-Functional Protein in Peroxisomal Beta-Oxidation. Structure and Substrate Specificity of the Arabidopsis Thaliana Protein, Mfp2
J.Biol.Chem., 285, 2010
|
|
7MLG
 
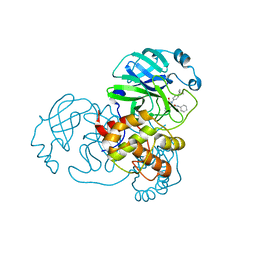 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) Covalently Bound to Compound C63 | | Descriptor: | (2R)-2-[(4-tert-butylphenyl)(ethanesulfonyl)amino]-N-cyclohexyl-2-(pyridin-3-yl)acetamide, 3C-like proteinase | | Authors: | Sharon, I, Stille, J, Tjutrins, J, Wang, G, Venegas, F.A, Hennecker, C, Rueda, A.M, Miron, C.E, Pinus, S, Labarre, A, Patrascu, M.B, Vlaho, D, Huot, M, Mittermaier, A.K, Moitessier, N, Schmeing, T.M. | | Deposit date: | 2021-04-28 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis and in vitro evaluation of novel SARS-CoV-2 3CL pro covalent inhibitors.
Eur.J.Med.Chem., 229, 2021
|
|
7MLF
 
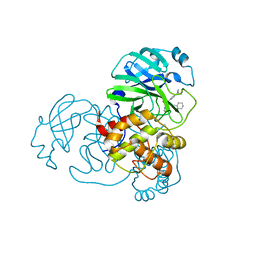 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) Covalently Bound to Compound C7 | | Descriptor: | 3C-like proteinase, N-(4-tert-butylphenyl)-2-chloro-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]acetamide | | Authors: | Sharon, I, Stille, J, Tjutrins, J, Wang, G, Venegas, F.A, Hennecker, C, Rueda, A.M, Miron, C.E, Pinus, S, Labarre, A, Patrascu, M.B, Vlaho, D, Huot, M, Mittermaier, A.K, Moitessier, N, Schmeing, T.M. | | Deposit date: | 2021-04-28 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis and in vitro evaluation of novel SARS-CoV-2 3CL pro covalent inhibitors.
Eur.J.Med.Chem., 229, 2021
|
|
7AQX
 
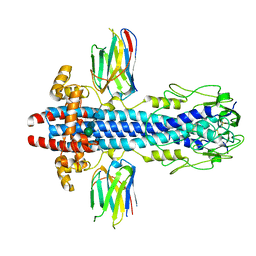 | |
7AQY
 
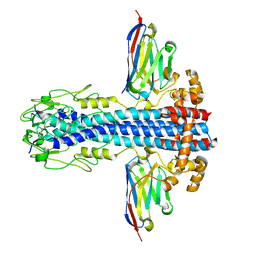 | |
7AR0
 
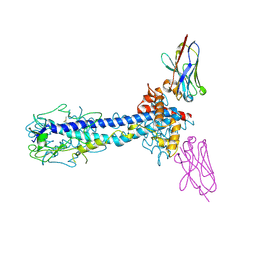 | |
7AQZ
 
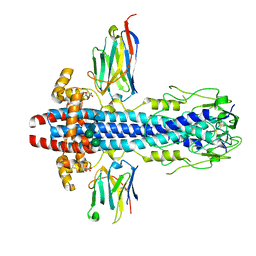 | | Co-Crystal Structure of Variant Surface Glycoprotein VSG2 in complex with Nanobody VSG2(NB14) | | Descriptor: | CITRIC ACID, Nanobody VSG2(NB14), SODIUM ION, ... | | Authors: | Stebbins, C.E, Hempelmann, A, VanStraaten, M. | | Deposit date: | 2020-10-23 | | Release date: | 2021-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Nanobody-mediated macromolecular crowding induces membrane fission and remodeling in the African trypanosome.
Cell Rep, 37, 2021
|
|
3IOQ
 
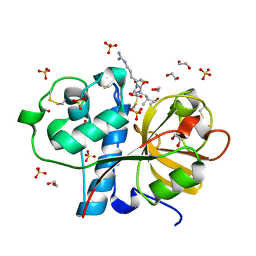 | | Crystal structure of the Carica candamarcensis cysteine protease CMS1MS2 in complex with E-64. | | Descriptor: | 1,2-ETHANEDIOL, CMS1MS2, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE, ... | | Authors: | Gomes, M.T.R, Teixeira, R.D, Salas, C.E, Nagem, R.A.P. | | Deposit date: | 2009-08-14 | | Release date: | 2010-02-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of the Carica candamarcensis cysteine protease CMS1MS2 in complex with E-64
To be Published
|
|
7BED
 
 | |
7BCY
 
 | |
280D
 
 | |
8EC3
 
 | |
1ULA
 
 | | APPLICATION OF CRYSTALLOGRAPHIC AND MODELING METHODS IN THE DESIGN OF PURINE NUCLEOSIDE PHOSPHORYLASE INHIBITORS | | Descriptor: | PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Ealick, S.E, Rule, S.A, Carter, D.C, Greenhough, T.J, Babu, Y.S, Cook, W.J, Habash, J, Helliwell, J.R, Stoeckler, J.D, Parksjunior, R.E, Chen, S.-F, Bugg, C.E. | | Deposit date: | 1991-11-05 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Application of crystallographic and modeling methods in the design of purine nucleoside phosphorylase inhibitors.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
1ULB
 
 | | APPLICATION OF CRYSTALLOGRAPHIC AND MODELING METHODS IN THE DESIGN OF PURINE NUCLEOSIDE PHOSPHORYLASE INHIBITORS | | Descriptor: | GUANINE, PURINE NUCLEOSIDE PHOSPHORYLASE, SULFATE ION | | Authors: | Ealick, S.E, Rule, S.A, Carter, D.C, Greenhough, T.J, Babu, Y.S, Cook, W.J, Habash, J, Helliwell, J.R, Stoeckler, J.D, Parksjunior, R.E, Chen, S.-F, Bugg, C.E. | | Deposit date: | 1991-11-05 | | Release date: | 1993-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Application of crystallographic and modeling methods in the design of purine nucleoside phosphorylase inhibitors.
Proc.Natl.Acad.Sci.USA, 88, 1991
|
|
