5TBY
 
 | | HUMAN BETA CARDIAC HEAVY MEROMYOSIN INTERACTING-HEADS MOTIF OBTAINED BY HOMOLOGY MODELING (USING SWISS-MODEL) OF HUMAN SEQUENCE FROM APHONOPELMA HOMOLOGY MODEL (PDB-3JBH), RIGIDLY FITTED TO HUMAN BETA-CARDIAC NEGATIVELY STAINED THICK FILAMENT 3D-RECONSTRUCTION (EMD-2240) | | Descriptor: | Myosin light chain 3, Myosin regulatory light chain 2, ventricular/cardiac muscle isoform, ... | | Authors: | ALAMO, L, WARE, J.S, PINTO, A, GILLILAN, R.E, SEIDMAN, J.G, SEIDMAN, C.E, PADRON, R. | | Deposit date: | 2016-09-13 | | Release date: | 2017-06-07 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes.
Elife, 6, 2017
|
|
5N28
 
 | | METHYL-COENZYME M REDUCTASE III FROM METHANOTORRIS FORMICICUS MONOCLINIC FORM | | Descriptor: | 1-THIOETHANESULFONIC ACID, Coenzyme B, FACTOR 430, ... | | Authors: | Wagner, T, Wegner, C.E, Ermler, U, Shima, S. | | Deposit date: | 2017-02-07 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phylogenetic and Structural Comparisons of the Three Types of Methyl Coenzyme M Reductase from Methanococcales and Methanobacteriales.
J.Bacteriol., 199, 2017
|
|
2N4L
 
 | | Solution Structure of the HIV-1 Intron Splicing Silencer and its Interactions with the UP1 Domain of hnRNP A1 | | Descriptor: | RNA (53-MER) | | Authors: | Tolbert, B.S, Jain, N, Morgan, C.E, Rife, B.D, Salemi, M. | | Deposit date: | 2015-06-23 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the HIV-1 Intron Splicing Silencer and Its Interactions with the UP1 Domain of Heterogeneous Nuclear Ribonucleoprotein (hnRNP) A1.
J.Biol.Chem., 291, 2016
|
|
5UVI
 
 | | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, ... | | Authors: | Martin-Garcia, J.M, Conrad, C.E, Nelson, G, Stander, N, Zatsepin, N.A, Zook, J, Zhu, L, Geiger, J, Chun, E, Kissick, D, Hilgart, M.C, Ogata, C, Ishchenko, A, Nagaratnam, N, Roy-Chowdhury, S, Coe, J, Subramanian, G, Schaffer, A, James, D, Ketawala, G, Venugopalan, N, Xu, S, Corcoran, S, Ferguson, D, Weierstall, U, Spence, J.C.H, Cherezov, V, Fromme, P, Fischetti, R.F, Liu, W. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation.
IUCrJ, 4, 2017
|
|
5NDZ
 
 | | Crystal structure of a thermostabilised human protease-activated receptor-2 (PAR2) in complex with AZ3451 at 3.6 angstrom resolution | | Descriptor: | 2-(6-bromanyl-1,3-benzodioxol-5-yl)-~{N}-(4-cyanophenyl)-1-[(1~{S})-1-cyclohexylethyl]benzimidazole-5-carboxamide, Lysozyme,Proteinase-activated receptor 2,Soluble cytochrome b562,Proteinase-activated receptor 2, SODIUM ION | | Authors: | Cheng, R.K.Y, Fiez-Vandal, C, Schlenker, O, Edman, K, Aggeler, B, Brown, D.G, Brown, G, Cooke, R.M, Dumelin, C.E, Dore, A.S, Geschwindner, S, Grebner, C, Hermansson, N.-O, Jazayeri, A, Johansson, P, Leong, L, Prihandoko, R, Rappas, M, Soutter, H, Snijder, A, Sundstrom, L, Tehan, B, Thornton, P, Troast, D, Wiggin, G, Zhukov, A, Marshall, F.H, Dekker, N. | | Deposit date: | 2017-03-09 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insight into allosteric modulation of protease-activated receptor 2.
Nature, 545, 2017
|
|
2OD3
 
 | | Human thrombin chimera with human residues 184a, 186, 186a, 186b, 186c and 222 replaced by murine thrombin equivalents. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-phenylalanyl-N-[(2S,3S)-6-{[amino(iminio)methyl]amino}-1-chloro-2-hydroxyhexan-3-yl]-L-prolinamide, Thrombin heavy chain, ... | | Authors: | Marino, F, Chen, Z, Ergenekan, C.E, Bush, L.A, Mathews, F.S, Di Cera, E. | | Deposit date: | 2006-12-21 | | Release date: | 2007-04-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis of na+ activation mimicry in murine thrombin.
J.Biol.Chem., 282, 2007
|
|
5UVJ
 
 | | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Martin-Garcia, J.M, Conrad, C.E, Nelson, G, Stander, N, Zatsepin, N.A, Zook, J, Zhu, L, Geiger, J, Chun, E, Kissick, D, Hilgart, M.C, Ogata, C, Ishchenko, A, Nagaratnam, N, Roy-Chowdhury, S, Coe, J, Subramanian, G, Schaffer, A, James, D, Ketawala, G, Venugopalan, N, Xu, S, Corcoran, S, Ferguson, D, Weierstall, U, Spence, J.C.H, Cherezov, V, Fromme, P, Fischetti, R.F, Liu, W. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation.
IUCrJ, 4, 2017
|
|
2NUH
 
 | |
5UVK
 
 | | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation | | Descriptor: | C-phycocyanin alpha chain, C-phycocyanin beta chain, PHYCOCYANOBILIN | | Authors: | Martin-Garcia, J.M, Conrad, C.E, Nelson, G, Stander, N, Zatsepin, N.A, Zook, J, Zhu, L, Geiger, J, Chun, E, Kissick, D, Hilgart, M.C, Ogata, C, Ishchenko, A, Nagaratnam, N, Roy-Chowdhury, S, Coe, J, Subramanian, G, Schaffer, A, James, D, Ketawala, G, Venugopalan, N, Xu, S, Corcoran, S, Ferguson, D, Weierstall, U, Spence, J.C.H, Cherezov, V, Fromme, P, Fischetti, R.F, Liu, W. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation.
Iucrj, 4, 2017
|
|
5UVL
 
 | | Serial Millisecond Crystallography of Membrane and Soluble Protein Micro-crystals using Synchrotron Radiation | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Martin-Garcia, J.M, Conrad, C.E, Nelson, G, Stander, N, Zatsepin, N.A, Zook, J, Zhu, L, Geiger, J, Chun, E, Kissick, D, Hilgart, M.C, Ogata, C, Ishchenko, A, Nagaratnam, N, Roy-Chowdhury, S, Coe, J, Subramanian, G, Schaffer, A, James, D, Ketawala, G, Venugopalan, N, Xu, S, Corcoran, S, Ferguson, D, Weierstall, U, Spence, J.C.H, Cherezov, V, Fromme, P, Fischetti, R.F, Liu, W. | | Deposit date: | 2017-02-20 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation.
IUCrJ, 4, 2017
|
|
1EIB
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT D313A COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-02-25 | | Release date: | 2001-02-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
1HLU
 
 | | STRUCTURE OF BOVINE BETA-ACTIN-PROFILIN COMPLEX WITH ACTIN BOUND ATP PHOSPHATES SOLVENT ACCESSIBLE | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, BETA-ACTIN, CALCIUM ION, ... | | Authors: | Chik, J.K, Lindberg, U, Schutt, C.E. | | Deposit date: | 1997-05-30 | | Release date: | 1997-10-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The structure of an open state of beta-actin at 2.65 A resolution.
J.Mol.Biol., 263, 1996
|
|
2O8Z
 
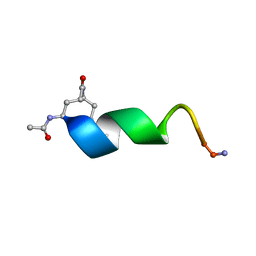 | | Bound Structure of CRF1 Extracellular Domain Antagonist | | Descriptor: | cCRF(30-41) Peptide | | Authors: | Mesleh, M.F, Shirley, W.A, Heise, C.E, Ling, N, Maki, R.A, Laura, R.P. | | Deposit date: | 2006-12-12 | | Release date: | 2006-12-26 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structural characterization of a minimal peptide antagonist bound to the extracellular domain of the corticotropin-releasing factor1 receptor.
J.Biol.Chem., 282, 2007
|
|
2M5N
 
 | | Atomic-resolution structure of a cross-beta protofilament | | Descriptor: | Transthyretin | | Authors: | Fitzpatrick, A.W.P, Debelouchina, G.T, Bayro, M.J, Clare, D.K, Caporini, M.A, Bajaj, V.S, Jaroniec, C.P, Wang, L, Ladizhansky, V, Muller, S, MacPhee, C.E, Waudby, C.A, Mott, H.R, de Simone, A, Knowles, T.P.J, Saibil, H.R, Vendruscolo, M, Orlova, E.V, Griffin, R.G, Dobson, C.M. | | Deposit date: | 2013-02-27 | | Release date: | 2013-07-17 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic structure and hierarchical assembly of a cross-{beta} amyloid fibril.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5VJH
 
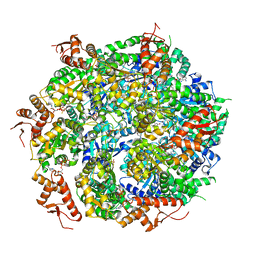 | | Closed State CryoEM Reconstruction of Hsp104:ATPyS and FITC casein | | Descriptor: | FITC casein, Heat shock protein 104, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2017-04-19 | | Release date: | 2017-07-05 | | Last modified: | 2017-08-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
2KQA
 
 | | The solution structure of the fungal elicitor Cerato-Platanin | | Descriptor: | Cerato-platanin | | Authors: | Oliveira, A.L, Gallo, M, Pazzagli, L, Cappugi, G, Scala, A, Cicero, D.O, Pantera, B, Spisni, A, Benedetti, C.E, Pertinhez, T.A. | | Deposit date: | 2009-11-03 | | Release date: | 2011-03-23 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the fungal elicitor Cerato-Platanin
To be Published
|
|
5VY8
 
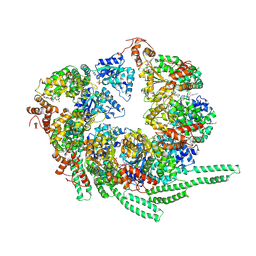 | | S. cerevisiae Hsp104-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein 104 | | Authors: | Gates, S.N, Yokom, A.L, Lin, J.-B, Jackrel, M.E, Rizo, A.N, Kendsersky, N.M, Buell, C.E, Sweeny, E.A, Chuang, E, Torrente, M.P, Mack, K.L, Su, M, Shorter, J, Southworth, D.R. | | Deposit date: | 2017-05-24 | | Release date: | 2017-07-05 | | Last modified: | 2018-08-15 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104.
Science, 357, 2017
|
|
2OCV
 
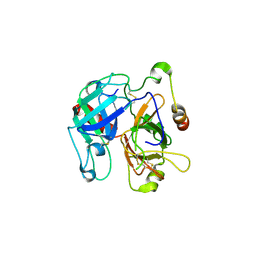 | | Structural basis of Na+ activation mimicry in murine thrombin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Thrombin | | Authors: | Marino, F, Chen, Z, Ergenekan, C.E, Bush, L.A, Mathews, F.S, Di Cera, E. | | Deposit date: | 2006-12-21 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of na+ activation mimicry in murine thrombin.
J.Biol.Chem., 282, 2007
|
|
1FFQ
 
 | | CRYSTAL STRUCTURE OF CHITINASE A COMPLEXED WITH ALLOSAMIDIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ALLOSAMIZOLINE, CHITINASE A | | Authors: | Papanikolau, Y, Tavlas, G, Vorgias, C.E, Petratos, K. | | Deposit date: | 2000-07-26 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | De novo purification scheme and crystallization conditions yield high-resolution structures of chitinase A and its complex with the inhibitor allosamidin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1EHN
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT E315Q COMPLEXED WITH OCTA-N-ACETYLCHITOOCTAOSE (NAG)8. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-02-22 | | Release date: | 2001-02-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
2OPR
 
 | | Crystal Structure of K101E Mutant HIV-1 Reverse Transcriptase in Complex with GW420867X. | | Descriptor: | ISOPROPYL (2S)-2-ETHYL-7-FLUORO-3-OXO-3,4-DIHYDROQUINOXALINE-1(2H)-CARBOXYLATE, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Weaver, K.L, Short, S.A, Chan, J.H, Kleim, J, Stammers, D.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Relationship of Potency and Resilience to Drug Resistant Mutations for GW420867X Revealed by Crystal Structures of Inhibitor Complexes for Wild-Type, Leu100Ile, Lys101Glu, and Tyr188Cys Mutant HIV-1 Reverse Transcriptases.
J.Med.Chem., 50, 2007
|
|
2OPS
 
 | | Crystal Structure of Y188C Mutant HIV-1 Reverse Transcriptase in Complex with GW420867X. | | Descriptor: | ISOPROPYL (2S)-2-ETHYL-7-FLUORO-3-OXO-3,4-DIHYDROQUINOXALINE-1(2H)-CARBOXYLATE, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Weaver, K.L, Short, S.A, Chan, J.H, Kleim, J, Stammers, D.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Relationship of Potency and Resilience to Drug Resistant Mutations for GW420867X Revealed by Crystal Structures of Inhibitor Complexes for Wild-Type, Leu100Ile, Lys101Glu, and Tyr188Cys Mutant HIV-1 Reverse Transcriptases.
J.Med.Chem., 50, 2007
|
|
2KQ5
 
 | |
2OPQ
 
 | | Crystal Structure of L100I Mutant HIV-1 Reverse Transcriptase in Complex with GW420867X. | | Descriptor: | ISOPROPYL (2S)-2-ETHYL-7-FLUORO-3-OXO-3,4-DIHYDROQUINOXALINE-1(2H)-CARBOXYLATE, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Weaver, K.L, Short, S.A, Chan, J.H, Kleim, J, Stammers, D.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Relationship of Potency and Resilience to Drug Resistant Mutations for GW420867X Revealed by Crystal Structures of Inhibitor Complexes for Wild-Type, Leu100Ile, Lys101Glu, and Tyr188Cys Mutant HIV-1 Reverse Transcriptases.
J.Med.Chem., 50, 2007
|
|
1FFR
 
 | | CRYSTAL STRUCTURE OF CHITINASE A MUTANT Y390F COMPLEXED WITH HEXA-N-ACETYLCHITOHEXAOSE (NAG)6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHITINASE A | | Authors: | Papanikolau, Y, Prag, G, Tavlas, G, Vorgias, C.E, Oppenheim, A.B, Petratos, K. | | Deposit date: | 2000-07-26 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High resolution structural analyses of mutant chitinase A complexes with substrates provide new insight into the mechanism of catalysis.
Biochemistry, 40, 2001
|
|
