5T4S
 
 | | Novel Approach of Fragment-Based Lead Discovery applied to Renin Inhibitors | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-chloro-N-[(furan-2-yl)methyl]pyrazin-2-amine, ... | | Authors: | Snell, G.P, Behnke, C.A, Okada, K, Oki, H, Sang, B.C, Lane, W. | | Deposit date: | 2016-08-30 | | Release date: | 2016-10-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Novel approach of fragment-based lead discovery applied to renin inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
5SY3
 
 | | Structure-based design of a new series of N-piperidin-3-ylpyrimidine-5-carboxamides as renin inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Snell, G.P, Behnke, C.A, Okada, K, Hideyuki, O, Sang, B.C, Lane, W. | | Deposit date: | 2016-08-10 | | Release date: | 2016-11-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-based design of a new series of N-(piperidin-3-yl)pyrimidine-5-carboxamides as renin inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
5TEI
 
 | | Structure of human ALDH1A1 with inhibitor CM039 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 6-{[(3-fluorophenyl)methyl]sulfanyl}-5-(2-methylphenyl)-2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, ... | | Authors: | Morgan, C.A, Hurley, T.D. | | Deposit date: | 2016-09-21 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of human ALDH1A1 with inhibitor CM039
To Be Published
|
|
5SZ9
 
 | | Structure-based design of a new series of N-piperidin-3-ylpyrimidine-5-carboxamides as renin inhibitors | | Descriptor: | (azepan-1-yl)(2-{[(furan-2-yl)methyl]amino}-6-methylpyridin-3-yl)methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Snell, G.P, Behnke, C.A, Okada, K, Hideyuki, O, Sang, B.C, Lane, W. | | Deposit date: | 2016-08-12 | | Release date: | 2016-11-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based design of a new series of N-(piperidin-3-yl)pyrimidine-5-carboxamides as renin inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
5SXN
 
 | | Structure-based design of a new series of N-piperidin-3-ylpyrimidine-5-carboxamides as renin inhibitors | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Snell, G.P, Behnke, C.A, Okada, K, Hideyuki, O, Sang, B.-C, Lane, W. | | Deposit date: | 2016-08-09 | | Release date: | 2016-11-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design of a new series of N-(piperidin-3-yl)pyrimidine-5-carboxamides as renin inhibitors.
Bioorg.Med.Chem., 24, 2016
|
|
5T0V
 
 | | Architecture of the Yeast Mitochondrial Iron-Sulfur Cluster Assembly Machinery: the Sub-Complex Formed by the Iron Donor, Yfh1, and the Scaffold, Isu1 | | Descriptor: | Frataxin homolog, mitochondrial, Iron sulfur cluster assembly protein 1 | | Authors: | Ranatunga, W, Gakh, O, Galeano, B.K, Smith IV, D.Y, Soderberg, C.A, Al-Karadaghi, S, Thompson, J.R, Isaya, G. | | Deposit date: | 2016-08-16 | | Release date: | 2016-08-31 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (17.5 Å) | | Cite: | Architecture of the Yeast Mitochondrial Iron-Sulfur Cluster Assembly Machinery: the Sub-Complex Formed by the Iron Donor, Yfh1, and the Scaffold, Isu1
J. Biol. Chem., 291, 2016
|
|
5T97
 
 | | ESTROGEN RECEPTOR ALPHA LIGAND BINDING DOMAIN IN COMPLEX WITH (2E)-3-(4-{(1R)-6-hydroxy-1-methyl-2-[4-(propan-2 -yl)phenyl]-1,2,3,4- tetrahydroisoquinolin-1-yl}phenyl)prop-2-enoic acid | | Descriptor: | (2E)-3-(4-{(1R)-6-hydroxy-1-methyl-2-[4-(propan-2-yl)phenyl]-1,2,3,4-tetrahydroisoquinolin-1-yl}phenyl)prop-2-enoic acid, Estrogen receptor | | Authors: | Kirby, C.A, Baird, J. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery of an Acrylic Acid Based Tetrahydroisoquinoline as an Orally Bioavailable Selective Estrogen Receptor Degrader for ER alpha + Breast Cancer.
J. Med. Chem., 60, 2017
|
|
1CJG
 
 | | NMR STRUCTURE OF LAC REPRESSOR HP62-DNA COMPLEX | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3'), PROTEIN (LAC REPRESSOR) | | Authors: | Spronk, C.A.E.M, Bonvin, A.M.J.J, Radha, P.K, Melacini, G, Boelens, R, Kaptein, R. | | Deposit date: | 1999-04-14 | | Release date: | 2000-01-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of Lac repressor headpiece 62 complexed to a symmetrical lac operator.
Structure Fold.Des., 7, 1999
|
|
1XB0
 
 | | Structure of the BIR domain of IAP-like protein 2 | | Descriptor: | Baculoviral IAP repeat-containing protein 8, Diablo homolog, mitochondrial, ... | | Authors: | Shin, H, Renatus, M, Eckelman, B.P, Nunes, V.A, Sampaio, C.A.M, Salvesen, G.S. | | Deposit date: | 2004-08-27 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The BIR domain of IAP-like protein 2 is conformationally unstable: implications for caspase inhibition
Biochem.J., 385, 2005
|
|
1XB1
 
 | | The Structure of the BIR domain of IAP-like protein 2 | | Descriptor: | Baculoviral IAP repeat-containing protein 8, Diablo homolog, mitochondrial, ... | | Authors: | Shin, H, Renatus, M, Eckelman, B.P, Nunes, V.A, Sampaio, C.A.M, Salvesen, G.S. | | Deposit date: | 2004-08-27 | | Release date: | 2004-11-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The BIR domain of IAP-like protein 2 is conformationally unstable: implications for caspase inhibition
Biochem.J., 385, 2005
|
|
3ZCX
 
 | |
3ZS5
 
 | | Structural basis for kinase selectivity of three clinical p38alpha inhibitors | | Descriptor: | 1,2-ETHANEDIOL, 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MITOGEN-ACTIVATED PROTEIN KINASE 14, ... | | Authors: | Azevedo, R, van Zeeland, M, Raaijmakers, H.C.A, Kazemier, B, Oubrie, A. | | Deposit date: | 2011-06-23 | | Release date: | 2012-07-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray structure of p38 alpha bound to TAK-715: comparison with three classic inhibitors.
Acta Crystallogr. D Biol. Crystallogr., 68, 2012
|
|
1E7L
 
 | | Endonuclease VII (EndoVII) N62D mutant from phage T4 | | Descriptor: | RECOMBINATION ENDONUCLEASE VII, SULFATE ION, ZINC ION | | Authors: | Raaijmakers, H.C.A, Vix, O, Toro, I, Suck, D. | | Deposit date: | 2000-08-29 | | Release date: | 2001-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Conformational Flexibility in T4 Endonuclease Vii Revealed by Crystallography: Implications for Substrate Binding and Cleavage
J.Mol.Biol., 308, 2001
|
|
4AW6
 
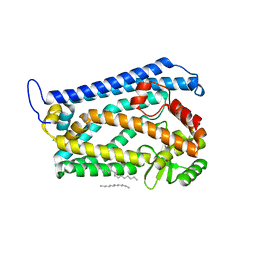 | | Crystal structure of the human nuclear membrane zinc metalloprotease ZMPSTE24 (FACE1) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CAAX PRENYL PROTEASE 1 HOMOLOG, ZINC ION | | Authors: | Pike, A.C.W, Dong, Y.Y, Quigley, A, Dong, L, Cooper, C.D.O, Chaikuad, A, Goubin, S, Shrestha, L, Li, Q, Mukhopadhyay, S, Yang, J, Xia, X, Shintre, C.A, Barr, A.J, Berridge, G, Chalk, R, Bray, J.E, von Delft, F, Bullock, A, Bountra, C, Arrowsmith, C.H, Edwards, A, Burgess-Brown, N, Carpenter, E.P. | | Deposit date: | 2012-05-31 | | Release date: | 2012-07-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Structural Basis of Zmpste24-Dependent Laminopathies.
Science, 339, 2013
|
|
4C3D
 
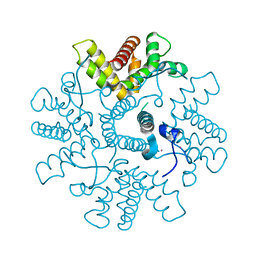 | | HRSV M2-1, P422 crystal form | | Descriptor: | CADMIUM ION, MATRIX M2-1 | | Authors: | Tanner, S.J, Ariza, A, Richard, C.A, Wu, W, Trincao, J, Hiscox, J.A, Carroll, M.W, Silman, N.J, Eleouet, J.F, Edwards, T.A, Barr, J.N. | | Deposit date: | 2013-08-22 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal Structure of the Essential Transcription Antiterminator M2-1 Protein of Human Respiratory Syncytial Virus and Implications of its Phosphorylation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3H98
 
 | | Crystal structure of HCV NS5b 1b with (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyrimidine derivative | | Descriptor: | GLYCEROL, N-{3-[5-hydroxy-8-(3-methylbutyl)-7-oxo-7,8-dihydroimidazo[1,2-a]pyrimidin-6-yl]-1,1-dioxido-4H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Wang, G, Lei, H, Wang, X, Das, D, Mackinnon, C, Montalbetti, C.A.G, Mears, R, Gai, X, Bailey, S, Ruhrmund, D, Hooi, L, Misialek, S, Rajagopalan, R, Cheng, R.K.Y, Barker, J.L, Felicetti, B, Stoycheva, A, Buckman, B, Kossen, K, Seiwert, S, Beigelmana, L. | | Deposit date: | 2009-04-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | HCV NS5B polymerase inhibitors 2: Synthesis and in vitro activity of (1,1-dioxo-2H-[1,2,4]benzothiadiazin-3-yl) azolo[1,5-a]pyridine and azolo[1,5-a]pyrimidine derivatives.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1E7D
 
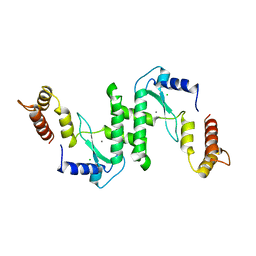 | | Endonuclease VII (ENDOVII) Ffrom Phage T4 | | Descriptor: | CALCIUM ION, CHLORIDE ION, RECOMBINATION ENDONUCLEASE VII, ... | | Authors: | Raaijmakers, H.C.A, Vix, O, Toro, I, Suck, D. | | Deposit date: | 2000-08-28 | | Release date: | 2001-05-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational Flexibility in T4 Endonuclease Vii Revealed by Crystallography: Implications for Substrate Binding and Cleavage
J.Mol.Biol., 308, 2001
|
|
4BW5
 
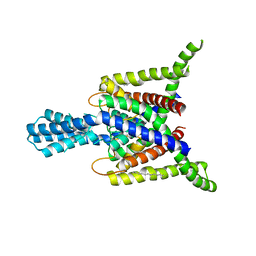 | | Crystal structure of human two pore domain potassium ion channel TREK2 (K2P10.1) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CADMIUM ION, POTASSIUM CHANNEL SUBFAMILY K MEMBER 10, ... | | Authors: | Pike, A.C.W, Dong, Y.Y, Dong, L, Quigley, A, Shrestha, L, Mukhopadhyay, S, Strain-Damerell, C, Goubin, S, Grieben, M, Shintre, C.A, Mackenzie, A, Vollmar, M, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Burgess-Brown, N, Carpenter, E.P. | | Deposit date: | 2013-06-30 | | Release date: | 2013-07-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | K2P Channel Gating Mechanisms Revealed by Structures of Trek-2 and a Complex with Prozac
Science, 347, 2015
|
|
3I03
 
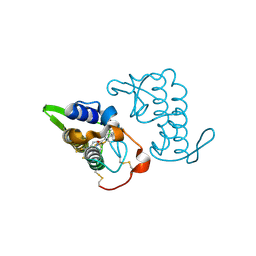 | | Crystal structure of bothropstoxin-I chemically modified by p-bromophenacyl bromide (BPB) - monomeric form at a high resolution | | Descriptor: | ISOPROPYL ALCOHOL, Phospholipase A2 homolog bothropstoxin-1, p-Bromophenacyl bromide | | Authors: | Marchi-Salvador, D.P, Fernandes, C.A.H, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
4C3B
 
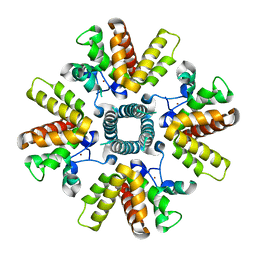 | | HRSV M2-1, P21 crystal form | | Descriptor: | MATRIX PROTEIN 2-1, ZINC ION | | Authors: | Tanner, S.J, Ariza, A, Richard, C.A, Wu, W, Trincao, J, Hiscox, J.A, Carroll, M.W, Silman, N.J, Eleouet, J.F, Edwards, T.A, Barr, J.N. | | Deposit date: | 2013-08-22 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Crystal Structure of the Essential Transcription Antiterminator M2-1 Protein of Human Respiratory Syncytial Virus and Implications of its Phosphorylation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4C3E
 
 | | HRSV M2-1 mutant S58D S61D, P21 crystal | | Descriptor: | MATRIX M2-1, ZINC ION | | Authors: | Tanner, S.J, Ariza, A, Richard, C.A, Wu, W, Trincao, J, Hiscox, J.A, Carroll, M.W, Silman, N.J, Eleouet, J.F, Edwards, T.A, Barr, J.N. | | Deposit date: | 2013-08-22 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Essential Transcription Antiterminator M2-1 Protein of Human Respiratory Syncytial Virus and Implications of its Phosphorylation.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3HZD
 
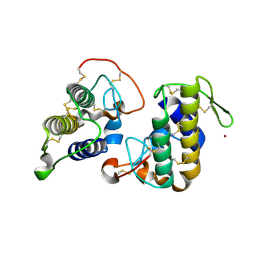 | | Crystal structure of bothropstoxin-I (BthTX-I), a PLA2 homologue from Bothrops jararacussu venom | | Descriptor: | LITHIUM ION, Phospholipase A2 homolog bothropstoxin-1 | | Authors: | Silva, M.C.O, Marchi-Salvador, D.P, Fernandes, C.A.H, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
4EC2
 
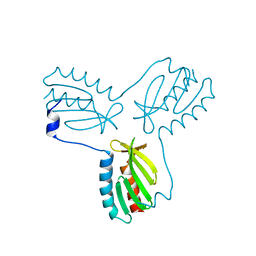 | | Crystal structure of trimeric frataxin from the yeast Saccharomyces cerevisiae, complexed with ferrous | | Descriptor: | FE (II) ION, Frataxin homolog, mitochondrial | | Authors: | Soderberg, C.A.G, Rajan, S, Gakh, O, Isaya, G, Al-Karadaghi, S. | | Deposit date: | 2012-03-26 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | The molecular basis of iron-induced oligomerization of frataxin and the role of the ferroxidation reaction in oligomerization.
J.Biol.Chem., 288, 2013
|
|
3HZW
 
 | | Crystal structure of bothropstoxin-I chemically modified by p-bromophenacyl bromide (BPB) | | Descriptor: | ISOPROPYL ALCOHOL, Phospholipase A2 homolog bothropstoxin-1, p-Bromophenacyl bromide | | Authors: | Fernandes, C.A.H, Marchi-Salvador, D.P, Soares, A.M, Fontes, M.R.M. | | Deposit date: | 2009-06-24 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Comparison between apo and complexed structures of bothropstoxin-I reveals the role of Lys122 and Ca(2+)-binding loop region for the catalytically inactive Lys49-PLA(2)s.
J.Struct.Biol., 171, 2010
|
|
4BB4
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, MAGNESIUM ION, N-(2-methoxyethyl)-4-[(6-pyridin-4-ylquinazolin-2-yl)amino]benzamide | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Optimization of a Novel Series of Potent Mutant B-Raf V600E Selective Kinase Inhibitors.
J.Med.Chem., 56, 2013
|
|
