3NJ1
 
 | | X-ray crystal structure of the PYL2(V114I)-pyrabactin A complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, GLYCEROL, ... | | Authors: | Peterson, F.C, Burgie, E.S, Bingman, C.A, Volkman, B.F, Phillips Jr, G.N, Cutler, S.R, Jensen, D.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-06-16 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Structural basis for selective activation of ABA receptors.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1NQT
 
 | | Crystal structure of bovine Glutamate dehydrogenase-ADP complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glutamate dehydrogenase 1 | | Authors: | Banerjee, S, Schmidt, T, Fang, J, Stanley, C.A, Smith, T.J. | | Deposit date: | 2003-01-23 | | Release date: | 2003-05-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural studies on ADP activation of mammalian glutamate dehydrogenase and the evolution of regulation
Biochemistry, 42, 2003
|
|
2FZ8
 
 | | Human Aldose reductase complexed with inhibitor zopolrestat at 1.48 A(1 day soaking). | | Descriptor: | 3,4-DIHYDRO-4-OXO-3-((5-TRIFLUOROMETHYL-2-BENZOTHIAZOLYL)METHYL)-1-PHTHALAZINE ACETIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, aldose reductase | | Authors: | Steuber, H, Zentgraf, M, Gerlach, C, Sotriffer, C.A, Heine, A, Klebe, G. | | Deposit date: | 2006-02-09 | | Release date: | 2006-10-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Expect the Unexpected or Caveat for Drug Designers: Multiple Structure Determinations Using Aldose Reductase Crystals Treated under Varying Soaking and Co-crystallisation Conditions.
J.Mol.Biol., 363, 2006
|
|
3NDY
 
 | |
2G0A
 
 | | X-ray structure of mouse pyrimidine 5'-nucleotidase type 1 with lead(II) bound in active site | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Cytosolic 5'-nucleotidase III, LEAD (II) ION | | Authors: | Bitto, E, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2006-02-11 | | Release date: | 2006-04-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of pyrimidine 5'-nucleotidase type 1. Insight into mechanism of action and inhibition during lead poisoning.
J.Biol.Chem., 281, 2006
|
|
4FZR
 
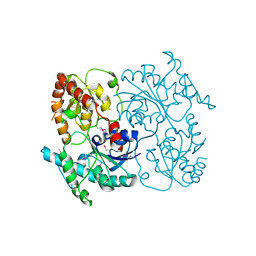 | | Crystal Structure of SsfS6, Streptomyces sp. SF2575 glycosyltransferase | | Descriptor: | SsfS6 | | Authors: | Wang, F, Zhou, M, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-07-07 | | Release date: | 2012-07-25 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Crystal structure of SsfS6, the putative C-glycosyltransferase involved in SF2575 biosynthesis.
Proteins, 81, 2013
|
|
3W87
 
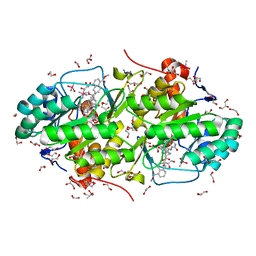 | | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with SH-1-103 | | Descriptor: | 1,2-ETHANEDIOL, 5-{4-[5-(methoxycarbonyl)naphthalen-2-yl]butyl}-2,6-dioxo-1,2,3,6-tetrahydropyrimidine-4-carboxylic acid, CACODYLATE ION, ... | | Authors: | Inaoka, D.K, Hashimoto, S, Rocha, J.R, Iida, M, Tabuchi, T, Lee, N, Matsuoka, S, Kuranaga, T, Shiba, T, Balogun, E.O, Sakamoto, K, Suzuki, S, Montanari, C.A, Nara, T, Aoki, T, Inoue, M, Honma, T, Tanaka, A, Harada, S, Kita, K. | | Deposit date: | 2013-03-12 | | Release date: | 2014-04-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structure of Trypanosoma cruzi dihydroorotate dehydrogenase in complex with SH-1-103
To be Published
|
|
6C2H
 
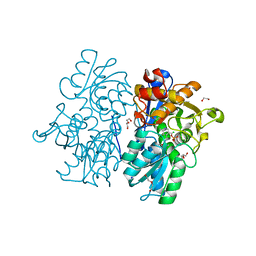 | | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: the Structure of the Catalytic Core | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Kreinbring, C.A, Tu, Y, Liu, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2018-01-08 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time.
Biochemistry, 57, 2018
|
|
3MNN
 
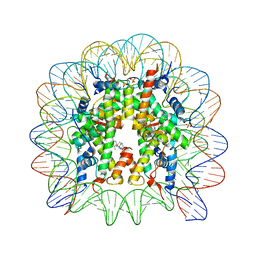 | | A Ruthenium Antitumour Agent Forms Specific Histone Protein Adducts in the Nucleosome Core | | Descriptor: | 1,3,5-triaza-7-phosphatricyclo[3.3.1.1~3,7~]decane, 1-methyl-4-(1-methylethyl)benzene, DNA (145-MER), ... | | Authors: | Ong, M.S, Davey, C.A. | | Deposit date: | 2010-04-22 | | Release date: | 2011-04-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A ruthenium antimetastasis agent forms specific histone protein adducts in the nucleosome core
Chemistry, 17, 2011
|
|
6C2N
 
 | | Crystal structure of HCV NS3/4A double mutant variant Y56H/D168A in complex with danoprevir | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, NS4A COFACTOR-NS3 PROTEIN CHIMERA, SULFATE ION, ... | | Authors: | Matthew, A.N, Schiffer, C.A. | | Deposit date: | 2018-01-08 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Clinical signature variant of HCV NS3/4A protease uses a novel mechanism to confer resistance
To Be Published
|
|
4G24
 
 | | Crystal Structure of proteinaceous RNase P 1 (PRORP1) from A. thaliana with Mn | | Descriptor: | 6-AMINOHEXANOIC ACID, MANGANESE (II) ION, Pentatricopeptide repeat-containing protein At2g32230, ... | | Authors: | Koutmos, M, Howard, M.J, Fierke, C.A. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mitochondrial ribonuclease P structure provides insight into the evolution of catalytic strategies for precursor-tRNA 5' processing.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1MFW
 
 | | STRUCTURE OF N-TERMINAL DOUBLECORTIN DOMAIN FROM DCLK: SELENOMETHIONINE LABELED PROTEIN | | Descriptor: | DOUBLECORTIN-LIKE KINASE (N-TERMINAL DOMAIN), SULFATE ION | | Authors: | Kim, M.H, Cierpickil, T, Derewenda, U, Krowarsch, D, Feng, Y, Devedjiev, Y, Dauter, Z, Walsh, C.A, Otlewski, J, Bushweller, J.H, Derewenda, Z. | | Deposit date: | 2002-08-13 | | Release date: | 2003-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The DCX-domain Tandems of Doublecortin and Doublecortin-like Kinase
Nat.Struct.Biol., 10, 2003
|
|
4G2T
 
 | | Crystal Structure of Streptomyces sp. SF2575 glycosyltransferase SsfS6, complexed with thymidine diphosphate | | Descriptor: | SsfS6, THYMIDINE-5'-DIPHOSPHATE | | Authors: | Wang, F, Zhou, M, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-07-12 | | Release date: | 2012-07-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Crystal structure of SsfS6, the putative C-glycosyltransferase involved in SF2575 biosynthesis.
Proteins, 81, 2013
|
|
6C4P
 
 | | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: the Structure of the PMP Complex | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, CALCIUM ION, ... | | Authors: | Kreinbring, C.A, Tu, Y, Liu, D, Berkowitz, D.B, Petsko, G.A, Ringe, D. | | Deposit date: | 2018-01-12 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time.
Biochemistry, 57, 2018
|
|
4JDQ
 
 | |
4HCY
 
 | | Structure of a eukaryotic thiaminase-I bound to the thiamin analogue 3-deazathiamin | | Descriptor: | 2-{4-[(4-amino-2-methylpyrimidin-5-yl)methyl]-3-methylthiophen-2-yl}ethanol, thiaminase-I | | Authors: | Kreinbring, C.A, Hubbard, P.A, Leeper, F.J, Hawksley, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2012-10-01 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of a eukaryotic thiaminase I.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6C6Q
 
 | |
1MHZ
 
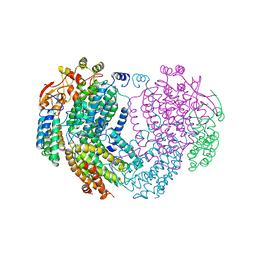 | | METHANE MONOOXYGENASE HYDROXYLASE | | Descriptor: | FE (III) ION, METHANE MONOOXYGENASE HYDROXYLASE | | Authors: | Elango, N, Radhakrishnan, R, Froland, W.A, Waller, B.J, Earhart, C.A, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1996-10-21 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b
Protein Sci., 6, 1997
|
|
3MXD
 
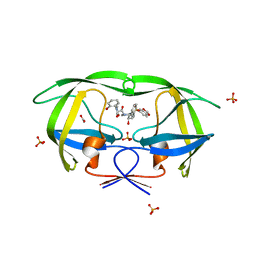 | | Crystal structure of HIV-1 protease inhibitor KC53 in complex with wild-type protease | | Descriptor: | (5S)-N-{(1S,2R)-3-[(1,3-benzodioxol-5-ylsulfonyl)(2-methylpropyl)amino]-1-benzyl-2-hydroxypropyl}-3-(2-hydroxyphenyl)-2 -oxo-1,3-oxazolidine-5-carboxamide, ACETATE ION, HIV-1 protease, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2010-05-07 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Design, Synthesis, and Structure-Activity Relationship Studies of HIV-1 Protease Inhibitors Incorporating Phenyloxazolidinones.
J.Med.Chem., 53, 2010
|
|
6C2Q
 
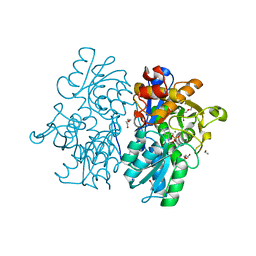 | | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: the Structure of the PLP-L-Serine Intermediate | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kreinbring, C.A, Tu, Y, Liu, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2018-01-08 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time.
Biochemistry, 57, 2018
|
|
6BV8
 
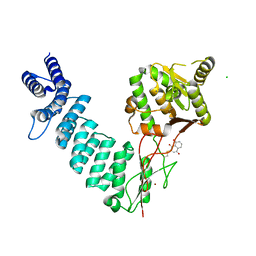 | | Structure of proteinaceous RNase P 1 (PRORP1) from A. thaliana complexed with Mn after 3-hour soak with juglone | | Descriptor: | 5-hydroxynaphthalene-1,4-dione, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Karasik, A, Wu, N, Fierke, C.A, Koutmos, M. | | Deposit date: | 2017-12-12 | | Release date: | 2019-06-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Inhibition of protein-only RNase P with Gambogic acid and Juglone
To Be Published
|
|
2GC5
 
 | | G51S mutant of L. casei FPGS | | Descriptor: | Folylpolyglutamate synthase, SULFATE ION | | Authors: | Smith, C.A, Cross, J.A, Bognar, A.L, Sun, X. | | Deposit date: | 2006-03-13 | | Release date: | 2006-06-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mutation of Gly51 to serine in the P-loop of Lactobacillus casei folylpolyglutamate synthetase abolishes activity by altering the conformation of two adjacent loops.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
4H9L
 
 | |
4LG0
 
 | | Structure of a ternary FOXO1-ETS1 DNA complex | | Descriptor: | CALCIUM ION, DNA (5'-D(*DAP*DAP*DAP*DCP*DAP*DAP*DTP*DAP*DAP*DCP*DAP*DGP*DGP*DAP*DAP*DAP*DCP*DCP*DGP*DTP*DG)-3'), DNA (5'-D(*DTP*DTP*DCP*DAP*DCP*DGP*DGP*DTP*DTP*DTP*DCP*DCP*DTP*DGP*DTP*DTP*DAP*DTP*DTP*DGP*DT)-3'), ... | | Authors: | Birrane, G, Choy, W.C, Datta, D, Geiger, C.A, Grant, M.A. | | Deposit date: | 2013-06-27 | | Release date: | 2014-07-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structure of a ternary FOXO1-ETS1 DNA complex
To be Published
|
|
2GC6
 
 | | S73A mutant of L. casei FPGS | | Descriptor: | Folylpolyglutamate synthase, SULFATE ION | | Authors: | Smith, C.A, Cross, J.A, Bognar, A.L, Sun, X. | | Deposit date: | 2006-03-13 | | Release date: | 2006-06-27 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mutation of Gly51 to serine in the P-loop of Lactobacillus casei folylpolyglutamate synthetase abolishes activity by altering the conformation of two adjacent loops.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
