5G5T
 
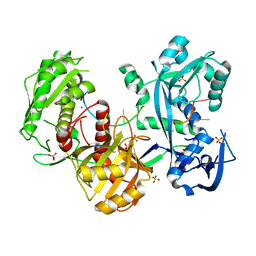 | | Structure of the Argonaute protein from Methanocaldcoccus janaschii in complex with guide DNA | | Descriptor: | ARGONAUTE, GUIDE DNA, MAGNESIUM ION, ... | | Authors: | Schneider, S, Oellig, C.A, Keegan, R, Grohmann, D, Zander, A, Willkomm, S. | | Deposit date: | 2016-06-03 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and mechanistic insights into an archaeal DNA-guided Argonaute protein.
Nat Microbiol, 2, 2017
|
|
1XF9
 
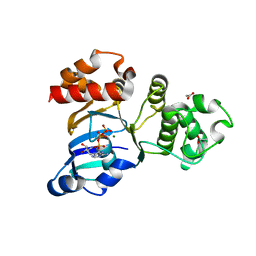 | | Structure of NBD1 from murine CFTR- F508S mutant | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, ... | | Authors: | Thibodeau, P.H, Brautigam, C.A, Machius, M, Thomas, P.J. | | Deposit date: | 2004-09-14 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Side chain and backbone contributions of Phe508 to CFTR folding.
Nat.Struct.Mol.Biol., 12, 2005
|
|
2XAO
 
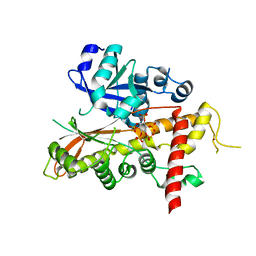 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with IP5 | | Descriptor: | INOSITOL-PENTAKISPHOSPHATE 2-KINASE, MYO-INOSITOL-(1,3,4,5,6)-PENTAKISPHOSPHATE, ZINC ION | | Authors: | Gonzalez, B, Banos-Sanz, J.I, Villate, M, Brearley, C.A, Sanz-Aparicio, J. | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase is a Distant Ipk Member with a Singular Inositide Binding Site for Axial 2-Oh Recognition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
1XDK
 
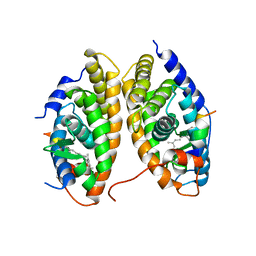 | | Crystal Structure of the RARbeta/RXRalpha Ligand Binding Domain Heterodimer in Complex with 9-cis Retinoic Acid and a Fragment of the TRAP220 Coactivator | | Descriptor: | (9cis)-retinoic acid, Retinoic acid receptor RXR-alpha, Retinoic acid receptor, ... | | Authors: | Pogenberg, V, Guichou, J.F, Vivat-Hannah, V, Kammerer, S, Perez, E, Germain, P, De Lera, A.R, Gronemeyer, H, Royer, C.A, Bourguet, W. | | Deposit date: | 2004-09-07 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | CHARACTERIZATION OF THE INTERACTION BETWEEN RAR/RXR HETERODIMERS AND TRANSCRIPTIONAL COACTIVATORS THROUGH STRUCTURAL AND FLUORESCENCE ANISOTROPY STUDIES
J.Biol.Chem., 280, 2005
|
|
5HOQ
 
 | | Apo structure of CalS11, TDP-rhamnose 3'-o-methyltransferase, an enzyme in Calicheamicin biosynthesis | | Descriptor: | SULFATE ION, TDP-rhamnose 3'-O-methyltransferase (CalS11) | | Authors: | Han, L, Helmich, K.E, Singh, S, Thorson, J.S, Bingman, C.A, Phillips Jr, G.N, Enzyme Discovery for Natural Product Biosynthesis | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Loop dynamics of thymidine diphosphate-rhamnose 3'-O-methyltransferase (CalS11), an enzyme in calicheamicin biosynthesis.
Struct Dyn., 3, 2016
|
|
1XMT
 
 | | X-ray structure of gene product from arabidopsis thaliana at1g77540 | | Descriptor: | BROMIDE ION, putative acetyltransferase | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-10-04 | | Release date: | 2004-10-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure of Arabidopsis thaliana At1g77540 Protein, a Minimal Acetyltransferase from the COG2388 Family.
Biochemistry, 45, 2006
|
|
1VY4
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in the pre-attack state of peptide bond formation containing acylated tRNA-substrates in the A and P sites. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Steitz, T.A, Innis, C.A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1W3K
 
 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN COMPLEX WITH CELLOBIO DERIVED-TETRAHYDROOXAZINE | | Descriptor: | ENDOGLUCANASE 5A, GLYCEROL, SULFATE ION, ... | | Authors: | Gloster, T.M, Macdonald, J.M, Tarling, C.A, Stick, R.V, Withers, S.W, Davies, G.J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural, Thermodynamic, and Kinetic Analyses of Tetrahydrooxazine-Derived Inhibitors Bound to {Beta}-Glucosidases
J.Biol.Chem., 279, 2004
|
|
1W3L
 
 | | ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN COMPLEX WITH CELLOTRI DERIVED-TETRAHYDROOXAZINE | | Descriptor: | ENDOGLUCANASE 5A, GLYCEROL, SULFATE ION, ... | | Authors: | Gloster, T.M, Macdonald, J.M, Tarling, C.A, Stick, R.V, Withers, S.W, Davies, G.J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Structural, Thermodynamic, and Kinetic Analyses of Tetrahydrooxazine-Derived Inhibitors Bound to {Beta}-Glucosidases
J.Biol.Chem., 279, 2004
|
|
1VY7
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in the pre-attack state of peptide bond formation containing short substrate-mimic Cytidine-Cytidine-Puromycin in the A site and acylated tRNA in the P site. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Steitz, T.A, Innis, C.A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome.
Nat.Struct.Mol.Biol., 21, 2014
|
|
5HGR
 
 | | Structure of Anabaena (Nostoc) sp. PCC 7120 Orange Carotenoid Protein binding canthaxanthin | | Descriptor: | Orange Carotenoid Protein (OCP), beta,beta-carotene-4,4'-dione | | Authors: | Sutter, M, Leverenz, R.L, Kerfeld, C.A. | | Deposit date: | 2016-01-08 | | Release date: | 2016-11-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.681 Å) | | Cite: | Different Functions of the Paralogs to the N-Terminal Domain of the Orange Carotenoid Protein in the Cyanobacterium Anabaena sp. PCC 7120.
Plant Physiol., 171, 2016
|
|
1VY5
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in the post-catalysis state of peptide bond formation containing dipeptydil-tRNA in the A site and deacylated tRNA in the P site. | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Polikanov, Y.S, Steitz, T.A, Innis, C.A. | | Deposit date: | 2014-05-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome.
Nat.Struct.Mol.Biol., 21, 2014
|
|
2XRP
 
 | | Human Doublecortin N-DC Repeat (1MJD) and Mammalian Tubulin (1JFF and 3HKE) Docked into the 8-Angstrom Cryo-EM Map of Doublecortin- Stabilised Microtubules | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, NEURONAL MIGRATION PROTEIN DOUBLECORTIN, ... | | Authors: | Fourniol, F.J, Sindelar, C.V, Amigues, B, Clare, D.K, Thomas, G, Perderiset, M, Francis, F, Houdusse, A, Moores, C.A. | | Deposit date: | 2010-09-18 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | Template-Free 13-Protofilament Microtubule-Map Assembly Visualized at 8 A Resolution.
J.Cell Biol., 191, 2010
|
|
1VM9
 
 | | The X-ray Structure of the cys84ala cys85ala double mutant of the [2Fe-2S] Ferredoxin subunit of Toluene-4-Monooxygenase from Pseudomonas Mendocina KR1 | | Descriptor: | 1,2-ETHANEDIOL, FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION, ... | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bingman, C.A, Allard, S.T.M, Moe, L.A, Fox, B.G, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-09-13 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure of T4moC, the Rieske-type ferredoxin component of toluene 4-monooxygenase.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5FT2
 
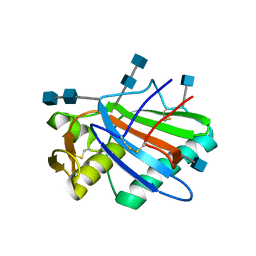 | | Sub-tomogram averaging of Lassa virus glycoprotein spike from virus- like particles at pH 5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PRE-GLYCOPROTEIN POLYPROTEIN GP COMPLEX | | Authors: | Li, S, Zhaoyang, S, Pryce, R, Parsy, M.L, Fehling, S.K, Schlie, K, Siebert, C.A, Garten, W, Bowden, T.A, Strecker, T, Huiskonen, J.T. | | Deposit date: | 2016-01-09 | | Release date: | 2016-03-02 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (16.4 Å) | | Cite: | Acidic Ph-Induced Conformations and Lamp1 Binding of the Lassa Virus Glycoprotein Spike.
Plos Pathog., 12, 2016
|
|
5FDU
 
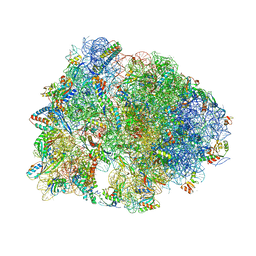 | | Crystal structure of the Metalnikowin I antimicrobial peptide bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Seefeldt, A.C, Graf, M, Perebaskine, N, Nguyen, F, Arenz, S, Mardirossian, M, Scocchi, M, Wilson, D.N, Innis, C.A. | | Deposit date: | 2015-12-16 | | Release date: | 2016-01-27 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the mammalian antimicrobial peptide Bac7(1-16) bound within the exit tunnel of a bacterial ribosome.
Nucleic Acids Res., 44, 2016
|
|
7TVL
 
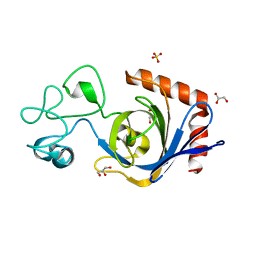 | | Viral AMG chitosanase V-Csn, apo structure | | Descriptor: | GLYCEROL, SULFATE ION, Viral chitosanase V-Csn | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
7TVN
 
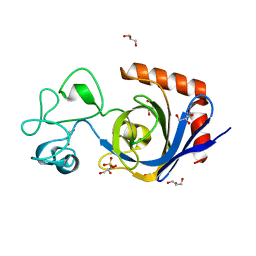 | | Viral AMG chitosanase V-Csn, D148N mutant | | Descriptor: | GLYCEROL, SULFATE ION, Viral chitosanase V-Csn D148N mutant | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
7TVO
 
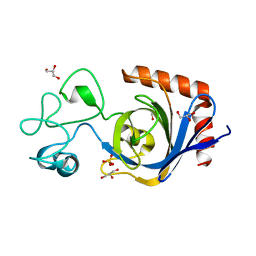 | | Viral AMG chitosanase V-Csn, E157Q mutant | | Descriptor: | GLYCEROL, SULFATE ION, Viral chitosanase V-Csn E157Q mutant | | Authors: | Smith, C.A, Wu, R, Buchko, G.W, Cort, J.R, Hofmockel, K.S, Jansson, J.K. | | Deposit date: | 2022-02-05 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural characterization of a soil viral auxiliary metabolic gene product - a functional chitosanase.
Nat Commun, 13, 2022
|
|
1VTT
 
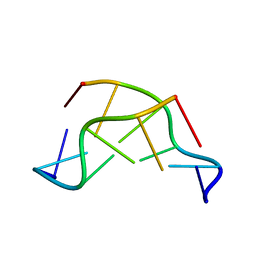 | | GT Wobble Base-Pairing in Z-DNA at 1.0 Angstrom Atomic Resolution: The Crystal Structure of d(CGCGTG) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*G)-3') | | Authors: | Ho, P.S, Frederick, C.A, Quigley, G.J, Van Der Marel, G.A, Van Boom, J.H, Wang, A.H.-J, Rich, A. | | Deposit date: | 1988-08-18 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | GT Wobble Base-Pairing in Z-DNA at 1.0 Angstrom Atomic Resolution: The Crystal Structure of d(CGCGTG)
Embo J., 4, 1985
|
|
5FET
 
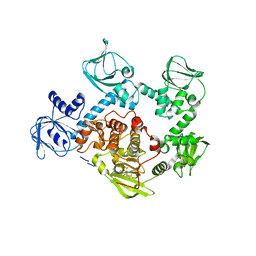 | | Crystal Structure of PVX_084705 in presence of Compound 2 | | Descriptor: | 4-[7-[(dimethylamino)methyl]-2-(4-fluorophenyl)imidazo[1,2-a]pyridin-3-yl]pyrimidin-2-amine, cGMP-dependent protein kinase, putative | | Authors: | Wernimont, A.K, Tempel, W, Walker, J.R, He, H, Seitova, A, Hills, T, Neculai, A.M, Baker, D.A, Flueck, C, Kettleborough, C.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Hui, R, Hutchinson, A, El Bakkouri, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-12-17 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Crystal Structure of PVX_084705 in presence of Compound 2
To be published
|
|
5F83
 
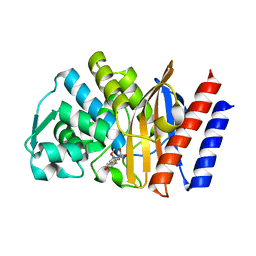 | | Imipenem complex of the GES-5 C69G mutant | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2015-12-09 | | Release date: | 2016-09-07 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Role of the Conserved Disulfide Bridge in Class A Carbapenemases.
J.Biol.Chem., 291, 2016
|
|
5F8K
 
 | | Crystal structure of the Bac7(1-16) antimicrobial peptide bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Seefeldt, A.C, Graf, M, Perebaskine, N, Nguyen, F, Arenz, S, Mardirossian, M, Scocchi, M, Wilson, D.N, Innis, C.A. | | Deposit date: | 2015-12-09 | | Release date: | 2016-02-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the mammalian antimicrobial peptide Bac7(1-16) bound within the exit tunnel of a bacterial ribosome.
Nucleic Acids Res., 44, 2016
|
|
1W9J
 
 | | Myosin II Dictyostelium discoideum motor domain S456Y bound with MgADP-AlF4 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Morris, C.A, Coureux, P.-D, Wells, A.L, Houdusse, A, Sweeney, H.L. | | Deposit date: | 2004-10-13 | | Release date: | 2006-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Function Analysis of Myosin II Backdoor Mutants
To be Published
|
|
1W9L
 
 | | Myosin II Dictyostelium discoideum motor domain S456E bound with MgADP-AlF4 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Morris, C.A, Coureux, P.-D, Wells, A.L, Houdusse, A, Sweeney, H.L. | | Deposit date: | 2004-10-13 | | Release date: | 2006-03-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Function Analysis of Myosin II Backdoor Mutants
To be Published
|
|
