2ISG
 
 | | Botulinum Neurotoxin A Light Chain WT Crystal Form B | | Descriptor: | NICKEL (II) ION, Neurotoxin BoNT/A, ZINC ION | | Authors: | Brunger, A.T, Stegmann, C.M. | | Deposit date: | 2006-10-17 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of metalloprotease botulinum serotype A from a pseudo-peptide binding mode to a small molecule that is active in primary neurons.
J.Biol.Chem., 282, 2007
|
|
2ISH
 
 | | Botulinum Neurotoxin A Light Chain WT Crystal Form C | | Descriptor: | Neurotoxin BoNT/A, ZINC ION | | Authors: | Brunger, A.T, Stegmann, C.M. | | Deposit date: | 2006-10-17 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibition of metalloprotease botulinum serotype A from a pseudo-peptide binding mode to a small molecule that is active in primary neurons.
J.Biol.Chem., 282, 2007
|
|
2ISE
 
 | | Botulinum Neurotoxin A Light Chain WT Crystal Form A | | Descriptor: | Neurotoxin BoNT/A, ZINC ION | | Authors: | Brunger, A.T, Stegmann, C.M. | | Deposit date: | 2006-10-17 | | Release date: | 2006-11-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibition of metalloprotease botulinum serotype A from a pseudo-peptide binding mode to a small molecule that is active in primary neurons.
J.Biol.Chem., 282, 2007
|
|
4PNJ
 
 | | Recombinant Sperm Whale P6 Myoglobin Solved with Single Pulse Free Electron Laser Data | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Cohen, A, Gonzalez, A, Lam, W, Lyubimov, A, Sauter, N, Tsai, Y, Uervirojnangkoorn, M, Brunger, A, Soltis, M. | | Deposit date: | 2014-05-23 | | Release date: | 2014-11-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Goniometer-based femtosecond crystallography with X-ray free electron lasers.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5W5C
 
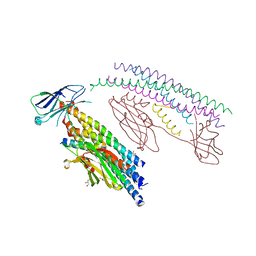 | |
6MDO
 
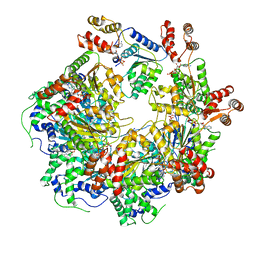 | | The D1 and D2 domain rings of NSF engaging the SNAP-25 N-terminus within the 20S supercomplex (focused refinement on D1/D2 rings, class 1) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Synaptosomal-associated protein 25, ... | | Authors: | White, K.I, Zhao, M, Brunger, A.T. | | Deposit date: | 2018-09-04 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural principles of SNARE complex recognition by the AAA+ protein NSF.
Elife, 7, 2018
|
|
7RZU
 
 | |
7RZS
 
 | |
7RZR
 
 | |
7RZT
 
 | |
7RZV
 
 | |
7RZQ
 
 | |
1N0Y
 
 | | Crystal Structure of Pb-bound Calmodulin | | Descriptor: | ACETATE ION, CACODYLATE ION, Calmodulin, ... | | Authors: | Wilson, M.A, Brunger, A.T. | | Deposit date: | 2002-10-15 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Domain flexibility in the 1.75 A resolution structure of Pb2+-calmodulin.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1EDU
 
 | | CRYSTAL STRUCTURE OF THE ENTH DOMAIN OF RAT EPSIN 1 | | Descriptor: | 1,2-ETHANEDIOL, EH domain binding protein EPSIN | | Authors: | Hyman, J.H, Chen, H, Decamilli, P, Brunger, A.T. | | Deposit date: | 2000-01-28 | | Release date: | 2000-05-10 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Epsin 1 undergoes nucleocytosolic shuttling and its eps15 interactor NH(2)-terminal homology (ENTH) domain, structurally similar to Armadillo and HEAT repeats, interacts with the transcription factor promyelocytic leukemia Zn(2)+ finger protein (PLZF).
J.Cell Biol., 149, 2000
|
|
3CF0
 
 | |
8CZI
 
 | |
7TIK
 
 | |
8FA2
 
 | |
8FA1
 
 | |
9BRR
 
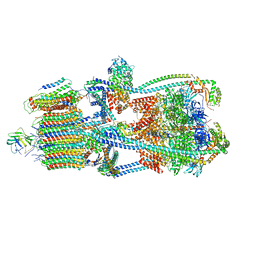 | | Intact V-ATPase State 3 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 2024
|
|
9BRA
 
 | | Intact V-ATPase State 2 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 2024
|
|
9BRQ
 
 | | Intact V-ATPase State 3 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 2024
|
|
9BRU
 
 | | Intact V-ATPase State 1 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 2024
|
|
9BRS
 
 | | Intact V-ATPase State 2 in synaptophysin knock-out isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, V-type proton ATPase 116 kDa subunit a 1, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 2024
|
|
9BRT
 
 | | Intact V-ATPase State 1 and synaptophysin complex in mouse brain isolated synaptic vesicles | | Descriptor: | Renin receptor cytoplasmic fragment, Ribonuclease kappa, Synaptophysin, ... | | Authors: | Wang, C, Jiang, W, Yang, K, Wang, X, Guo, Q, Brunger, A.T. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure and topography of the synaptic V-ATPase-synaptophysin complex.
Nature, 2024
|
|
