8GCR
 
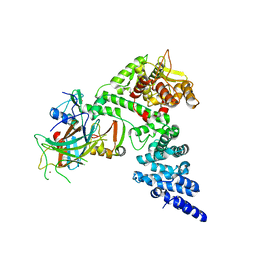 | | HPV16 E6-E6AP-p53 complex | | Descriptor: | Cellular tumor antigen p53, Maltose/maltodextrin-binding periplasmic protein,Protein E6, Ubiquitin-protein ligase E3A, ... | | Authors: | Bratkowski, M.A, Wang, J.C.K, Hao, Q, Nile, A.H. | | Deposit date: | 2023-03-02 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Structure of the p53 degradation complex from HPV16.
Nat Commun, 15, 2024
|
|
5BJS
 
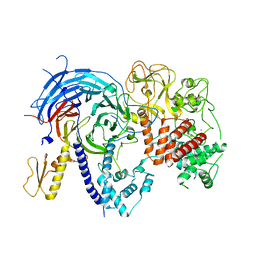 | | Apo ctPRC2 in an autoinhibited state | | Descriptor: | Histone-lysine N-methyltransferase EZH2, Polycomb protein SUZ12, Polycomb Protein EED, ... | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2016-10-22 | | Release date: | 2017-06-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.189 Å) | | Cite: | Polycomb repressive complex 2 in an autoinhibited state.
J. Biol. Chem., 292, 2017
|
|
8D0M
 
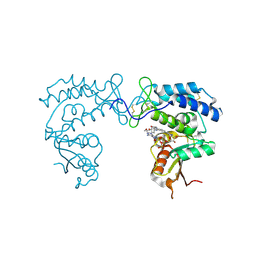 | | Human CD38 ectodomain bound to a 78c-ADPR adduct | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[5-[4-[[4-(2-methoxyethoxy)cyclohexyl]amino]-1-methyl-2-oxidanylidene-quinolin-6-yl]-1,3-thiazol-3-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Gu, W. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0I
 
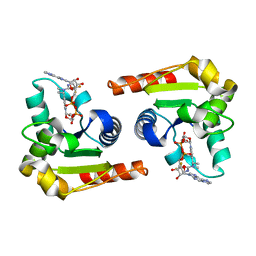 | | Human SARM1 bound to an NB-3 eADPR adduct | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-imidazo[2,1-f]purin-3-yl-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[4-[(1~{S})-1-[methyl-[2,2,2-tris(fluoranyl)ethylcarbamoyl]amino]ethyl]pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0C
 
 | | Human SARM1 TIR domain bound to NB-3-ADPR | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[4-[(1~{S})-1-[methyl-[2,2,2-tris(fluoranyl)ethylcarbamoyl]amino]ethyl]pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0G
 
 | | Human SARM1 TIR domain bound to NB-3-ADPRP | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{R},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3-oxidanyl-4-phosphonooxy-oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[4-[(1~{S})-1-[methyl-[2,2,2-tris(fluoranyl)ethylcarbamoyl]amino]ethyl]pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0D
 
 | | Human SARM1 TIR domain bound to an NB-7-ADPR adduct | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R})-5-[4-[3-[3-(4-chlorophenyl)propanoylamino]-4-methyl-1~{H}-pyrazol-5-yl]pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0H
 
 | | Human SARM1 TIR domain bound to NB-3-GDPR | | Descriptor: | NAD(+) hydrolase SARM1, [[(2~{R},3~{S},4~{R},5~{R})-5-(2-azanyl-6-oxidanylidene-3~{H}-purin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[4-[(1~{S})-1-[methyl-[2,2,2-tris(fluoranyl)ethylcarbamoyl]amino]ethyl]pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0F
 
 | | Human SARM1 TIR domain bound to NB-2-ADPR | | Descriptor: | NAD(+) hydrolase SARM1, [[(3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-[4-[[methyl-[2,2,2-tris(fluoranyl)ethylcarbamoyl]amino]methyl]pyridin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0E
 
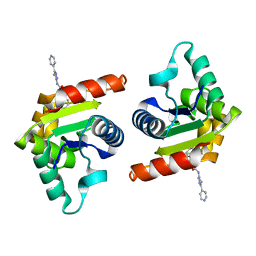 | | Human SARM1 TIR domain bound to NB-7 | | Descriptor: | 3-(4-chlorophenyl)-N-[4-methyl-3-(pyridin-4-yl)-1H-pyrazol-5-yl]propanamide, NAD(+) hydrolase SARM1 | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
8D0J
 
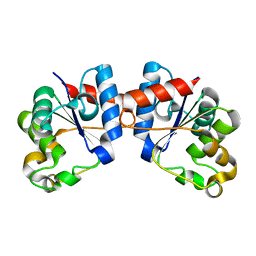 | | Apo Human SARM1 TIR domain | | Descriptor: | NAD(+) hydrolase SARM1 | | Authors: | Bratkowski, M.A, Mathur, P. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease.
Neuron, 110, 2022
|
|
5TQR
 
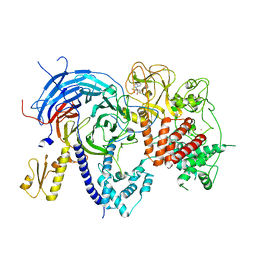 | |
5WG6
 
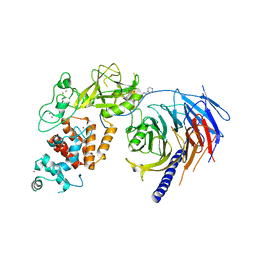 | | Human Polycomb Repressive Complex 2 in complex with GSK126 inhibitor | | Descriptor: | 1-[(2S)-butan-2-yl]-N-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3-methyl-6-[6-(piperazin-1-yl)pyridin-3-yl]-1H-indole-4-carboxamide, Histone-lysine N-methyltransferase EZH2,Polycomb protein SUZ12 (E.C.2.1.1.43) chimera, Polycomb protein EED, ... | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2017-07-13 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.901 Å) | | Cite: | An Evolutionarily Conserved Structural Platform for PRC2 Inhibition by a Class of Ezh2 Inhibitors.
Sci Rep, 8, 2018
|
|
5VK3
 
 | | Apo ctPRC2 with E840A and K852D mutations in Ezh2 | | Descriptor: | Histone-lysine N-methyltransferase EZH2,Polycomb protein SUZ12, Polycomb Protein EED, ZINC ION | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2017-04-20 | | Release date: | 2017-06-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.114 Å) | | Cite: | Polycomb repressive complex 2 in an autoinhibited state.
J. Biol. Chem., 292, 2017
|
|
5WFC
 
 | |
5WFD
 
 | | Humanized mutant of the Chaetomium thermophilum Polycomb Repressive Complex 2 bound to the inhibitor GSK126 | | Descriptor: | 1-[(2S)-butan-2-yl]-N-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3-methyl-6-[6-(piperazin-1-yl)pyridin-3-yl]-1H-indole-4-carboxamide, Histone-lysine-N-methyltransferase EZH2, Polycomb protein SUZ12 chimera, ... | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2017-07-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | An Evolutionarily Conserved Structural Platform for PRC2 Inhibition by a Class of Ezh2 Inhibitors.
Sci Rep, 8, 2018
|
|
5WH1
 
 | |
5WF7
 
 | | Chaetomium thermophilum Polycomb Repressive Complex 2 bound to GSK126 | | Descriptor: | 1-[(2S)-butan-2-yl]-N-[(4,6-dimethyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-3-methyl-6-[6-(piperazin-1-yl)pyridin-3-yl]-1H-indole-4-carboxamide, Histone-lysine-N-methyltransferase EZH2, Polycomb protein SUZ12 chimera, ... | | Authors: | Bratkowski, M.A, Liu, X. | | Deposit date: | 2017-07-11 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Evolutionarily Conserved Structural Platform for PRC2 Inhibition by a Class of Ezh2 Inhibitors.
Sci Rep, 8, 2018
|
|
8SL1
 
 | | Cryo-EM structure of PAPP-A2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Judge, R.A, Stoll, V.S, Eaton, D, Hao, Q, Bratkowski, M.A. | | Deposit date: | 2023-04-20 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Cryo-EM structure of human PAPP-A2 and mechanism of substrate recognition.
Commun Chem, 6, 2023
|
|
