5FBR
 
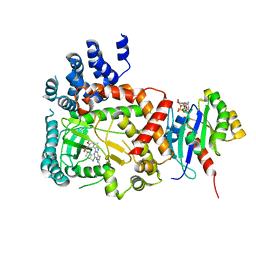 | | PI4KB in complex with Rab11 and the MI359 Inhibitor | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta, Ras-related protein Rab-11A, ... | | Authors: | Chalupska, D, Mejdrova, I, Nencka, R, Boura, E. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | PI4KB in complex with Rab11 and the MI359 Inhibitor
To Be Published
|
|
6HLW
 
 | |
6HLV
 
 | |
6HLT
 
 | |
6HMV
 
 | |
5FBL
 
 | | PI4KB in complex with Rab11 and the MI356 Inhibitor | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta, Ras-related protein Rab-11A, ... | | Authors: | Chalupska, D, Mejdrova, I, Nencka, R, Boura, E. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.372 Å) | | Cite: | PI4KB in complex with Rab11 and the MI356 Inhibitor
To Be Published
|
|
5FBV
 
 | | PI4KB in complex with Rab11 and the MI364 Inhibitor | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta, Ras-related protein Rab-11A, ... | | Authors: | Chalupska, D, Mejdrova, I, Nencka, R, Boura, E. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.285 Å) | | Cite: | PI4KB in complex with Rab11 and the MI364 Inhibitor
To Be Published
|
|
6HLN
 
 | |
5I14
 
 | | Truncated and mutated T4 lysozyme | | Descriptor: | NICKEL (II) ION, mutated and truncated T4 lysozyme | | Authors: | Klima, M, Boura, E. | | Deposit date: | 2016-02-05 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.745 Å) | | Cite: | Metal ions-binding T4 lysozyme as an intramolecular protein purification tag compatible with X-ray crystallography.
Protein Sci., 26, 2017
|
|
6S86
 
 | | Human wtSTING in complex with 3'3'-c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Stimulator of Interferon genes | | Authors: | Smola, M, Boura, E. | | Deposit date: | 2019-07-08 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | No magnesium is needed for binding of the stimulator of interferon genes to cyclic dinucleotides.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6S2A
 
 | | PI PLC mutant H82A | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, 1-phosphatidylinositol phosphodiesterase | | Authors: | Eisenreichova, A, Boura, E. | | Deposit date: | 2019-06-20 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | PI PLC mutant H82A
To Be Published
|
|
6S27
 
 | |
8A23
 
 | | Crystal structure of SARS-CoV-2 nsp10/nsp16 methyltransferase in complex with TO383 | | Descriptor: | (2R,3R,4S,5R)-2-[4-azanyl-5-(2-quinolin-3-ylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-5-(hydroxymethyl)oxolane-3,4-diol, 2'-O-methyltransferase nsp16, GLYCEROL, ... | | Authors: | Hanigovsky, M, Krafcikova, P, Klima, M, Boura, E. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of SARS-CoV-2 nsp10/nsp16 methyltransferase in
complex with TO383
To Be Published
|
|
7ZKU
 
 | | Crystal structure of human STING in complex with 3',3'-c-(2'F,2'dAMP-2'dGMP) | | Descriptor: | 9-[(1~{S},6~{R},8~{R},9~{R},10~{R},15~{R},17~{R})-8-(6-aminopurin-9-yl)-9-fluoranyl-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadecan-17-yl]-2-azanyl-3~{H}-purin-6-one, Stimulator of interferon protein | | Authors: | Klima, M, Smola, M, Boura, E. | | Deposit date: | 2022-04-13 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human STING in complex with 3',3'-c-(2'F,2'dAMP-2'dGMP)
To Be Published
|
|
7ZWL
 
 | | Crystal structure of human STING in complex with 3',3'-c-di-(2'F,2'dAMP) | | Descriptor: | 9-[(1~{R},6~{R},8~{R},9~{S},10~{R},15~{R},17~{R},18~{S})-17-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,11,13-tetraoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadecan-8-yl]purin-6-amine, Stimulator of interferon protein, Ubiquitin-like protein SMT3 | | Authors: | Klima, M, Smola, M, Boura, E. | | Deposit date: | 2022-05-19 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human STING in complex with 3',3'-c-di-(2'F,2'dAMP)
To Be Published
|
|
7ZV0
 
 | | Crystal structure of human STING in complex with 3',3'-c-(2'F,2'dAMP-2'F,2'dAMP) | | Descriptor: | 9-[(1~{R},6~{R},8~{R},9~{R},10~{R},15~{R},17~{R},18~{S})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadecan-17-yl]purin-6-amine, Stimulator of interferon protein | | Authors: | Klima, M, Smola, M, Boura, E. | | Deposit date: | 2022-05-13 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of human STING in complex with 3',3'-c-(2'F,2'dAMP-2'F,2'dAMP)
To Be Published
|
|
7ZVK
 
 | | Crystal structure of human STING in complex with 3',3'-c-(2'F,2'dAMP-IMP) | | Descriptor: | 9-[(1~{R},6~{R},8~{R},9~{R},10~{R},15~{R},17~{R},18~{S})-8-(6-aminopurin-9-yl)-9-fluoranyl-3,12,18-tris(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadecan-17-yl]-3~{H}-purin-6-one, Stimulator of interferon protein | | Authors: | Klima, M, Smola, M, Boura, E. | | Deposit date: | 2022-05-16 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Crystal structure of human STING in complex with 3',3'-c-(2'F,2'dAMP-IMP)
To Be Published
|
|
7ZXB
 
 | | Crystal structure of human STING in complex with 3',3'-c-(2'dAMP-2'F,2'dAMP) | | Descriptor: | 9-[(1~{R},6~{R},8~{R},10~{S},15~{R},17~{R},18~{S})-8-(6-aminopurin-9-yl)-18-fluoranyl-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadecan-17-yl]purin-6-amine, Stimulator of interferon protein | | Authors: | Klima, M, Smola, M, Boura, E. | | Deposit date: | 2022-05-20 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human STING in complex with 3',3'-c-(2'dAMP-2'F,2'dAMP)
To Be Published
|
|
8A2X
 
 | | Crystal structure of human STING in complex with 3',3'-c-(2'F,2'dAMP(S)-2'F,2'dAMP(S)) | | Descriptor: | 9-[(1~{R},3~{R},6~{R},8~{R},9~{R},10~{R},12~{R},15~{R},17~{R},18~{S})-8-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13-pentaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadecan-17-yl]purin-6-amine, Stimulator of interferon protein | | Authors: | Klima, M, Smola, M, Boura, E. | | Deposit date: | 2022-06-06 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of human STING in complex with 3',3'-c-(2'F,2'dAMP(S)-2'F,2'dAMP(S))
To Be Published
|
|
8B07
 
 | |
8A5X
 
 | |
7ZIT
 
 | | 14-3-3 in complex with SARS-COV2 N phospho-peptide | | Descriptor: | 14-3-3 protein zeta/delta, ACETATE ION, BENZOIC ACID, ... | | Authors: | Eisenreichova, A, Boura, E. | | Deposit date: | 2022-04-08 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural basis for SARS-CoV-2 nucleocapsid (N) protein recognition by 14-3-3 proteins.
J.Struct.Biol., 214, 2022
|
|
8A2H
 
 | | human STING in complex with 2',3'-cyclic-GMP-7-deazaphenyl-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-(4-azanyl-5-phenyl-pyrrolo[2,3-d]pyrimidin-7-yl)-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Smola, M, Vavrina, Z, Boura, E, Brynda, J. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design, Synthesis, and Biochemical and Biological Evaluation of Novel 7-Deazapurine Cyclic Dinucleotide Analogues as STING Receptor Agonists.
J.Med.Chem., 65, 2022
|
|
6TC6
 
 | |
6TC9
 
 | | Crystal structure of MutM from Neisseria meningitidis | | Descriptor: | DNA, DNA containing abasic site analogue, Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Silhan, J, Landova, B, Boura, E. | | Deposit date: | 2019-11-05 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.175 Å) | | Cite: | Conformational changes of DNA repair glycosylase MutM triggered by DNA binding.
Febs Lett., 594, 2020
|
|
