4UF0
 
 | | Crystal structure of JmjC domain of human histone demethylase UTY in complex with epitherapuetic compound 2-(((2-((2-(dimethylamino)ethyl) (ethyl)amino)-2-oxoethyl)amino)methyl)isonicotinic acid. | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(2-{[(E)-2-(dimethylamino)ethenyl](ethyl)amino}-2-oxoethyl)amino]methyl}pyridine-4-carboxylic acid, FE (II) ION, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Tobias, K, Kopec, J, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2014-12-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
4UN0
 
 | | Crystal structure of the human CDK12-cyclinK complex | | Descriptor: | CYCLIN-DEPENDENT KINASE 12, CYCLIN-K | | Authors: | Dixon Clarke, S.E, Elkins, J.M, Pike, A.C.W, Chaikuad, A, Goubin, S, Krojer, T, Sorrell, F.J, Nowak, R, Williams, E, Kopec, J, Mahajan, R.P, Burgess-Brown, N, Carpenter, E.P, Knapp, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-05-22 | | Release date: | 2014-06-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures of the Cdk12/Cyck Complex with AMP-Pnp Reveal a Flexible C-Terminal Kinase Extension Important for ATP Binding.
Sci.Rep., 5, 2015
|
|
5A15
 
 | | Crystal structure of the BTB domain of human KCTD16 | | Descriptor: | BTB/POZ DOMAIN-CONTAINING PROTEIN KCTD16 | | Authors: | Pinkas, D.M, Sanvitale, C.E, Solcan, N, Goubin, S, Canning, P, Dixon Clarke, S.E, Talon, R, Wiggers, H.J, Fitzpatrick, F, Tallant, C, Kopec, J, Chalk, R, Doutch, J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2015-04-28 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural complexity in the KCTD family of Cullin3-dependent E3 ubiquitin ligases.
Biochem. J., 474, 2017
|
|
6TTK
 
 | | Crystal structure of the kelch domain of human KLHL12 in complex with DVL1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DVL1, ... | | Authors: | Chen, Z, Williams, E, Pike, A.C.W, Strain-Damerell, C, Wang, D, Chalk, R, Burgess-Brown, N, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-12-27 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Identification of a PGXPP degron motif in dishevelled and structural basis for its binding to the E3 ligase KLHL12.
Open Biology, 10, 2020
|
|
5A7W
 
 | | Crystal structure of human JMJD2A in complex with compound 35 | | Descriptor: | 1,2-ETHANEDIOL, 2-[5-[(4-hydroxyphenyl)carbonylamino]-2-oxidanyl-phenyl]pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Kopec, J, Tallant, C, Froese, S, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | Deposit date: | 2015-07-10 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
5A7S
 
 | | Crystal structure of human JMJD2A in complex with compound 44 | | Descriptor: | 1,2-ETHANEDIOL, 2-(5-acetamido-2-oxidanyl-phenyl)pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Kopec, J, Tallant, C, Froese, S, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | Deposit date: | 2015-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
3H9U
 
 | | S-adenosyl homocysteine hydrolase (SAHH) from Trypanosoma brucei | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, GLYCEROL, ... | | Authors: | Siponen, M.I, Schutz, P, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Karlberg, T, Kragh Nielsen, T, Kotenyova, T, Kotzsch, A, Moche, M, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Wisniewska, M, Schueler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of S-adenosyl homocysteine hydrolase (SAHH) from Trypanosoma brucei
To be Published
|
|
6ES0
 
 | | Crystal structure of the kinase domain of human RIPK2 in complex with the activation loop targeting inhibitor CS-R35 | | Descriptor: | 2-[2-fluoranyl-4-[[2-fluoranyl-4-[2-(methylcarbamoyl)pyridin-4-yl]oxy-phenyl]carbamoylamino]phenyl]sulfanylethanoic acid, Receptor-interacting serine/threonine-protein kinase 2 | | Authors: | Pinkas, D.M, Bufton, J.C, Suebsuwong, C, Ray, S.S, Dai, B, Newman, J.A, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Degterev, A, Cuny, G.D, Bullock, A.N. | | Deposit date: | 2017-10-19 | | Release date: | 2018-02-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Activation loop targeting strategy for design of receptor-interacting protein kinase 2 (RIPK2) inhibitors.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
6EIX
 
 | | Crystal structure of the kinase domain of the Q207E mutant of ACVR1 (ALK2) in complex with a 2-aminopyridine inhibitor K02288 | | Descriptor: | 1,2-ETHANEDIOL, 3-[6-amino-5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenol, Activin receptor type-1 | | Authors: | Williams, E.P, Canning, P, Sanvitale, C.E, Krojer, T, Allerston, C.K, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2017-09-19 | | Release date: | 2017-09-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the kinase domain of the Q207E mutant of ACVR1 (ALK2) in complex with K02288
To be published
|
|
3IUC
 
 | | Crystal structure of the human 70kDa heat shock protein 5 (BiP/GRP78) ATPase domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, Heat shock 70kDa protein 5 (Glucose-regulated protein, ... | | Authors: | Wisniewska, M, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Kotyenova, T, Kotzch, A, Kraulis, P, Markova, N, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Roos, A, Schutz, P, Siponen, M.I, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-31 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B', and HSPA5/BiP/GRP78
Plos One, 5, 2010
|
|
6EQL
 
 | | Crystal Structure of Human Glycogenin-1 (GYG1) Tyr195pIPhe mutant complexed with manganese and UDP | | Descriptor: | 1,2-ETHANEDIOL, Glycogenin-1, MANGANESE (II) ION, ... | | Authors: | Bailey, H.J, Kopec, J, Bilyard, M.K, Bezerra, G.A, Seo Lee, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Davis, B.G, Yue, W.W. | | Deposit date: | 2017-10-13 | | Release date: | 2017-12-20 | | Last modified: | 2018-11-14 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Palladium-mediated enzyme activation suggests multiphase initiation of glycogenesis.
Nature, 563, 2018
|
|
4QC3
 
 | | Crystal structure of human BAZ2B bromodomain in complex with a diacetylated histone 4 peptide (H4K8acK12ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, diacetylated histone 4 peptide (H4K8acK12ac) | | Authors: | Tallant, C, Jose, B, Picaud, S, Chaikuad, A, Filippakopoulos, P, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-05-09 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular basis of histone tail recognition by human TIP5 PHD finger and bromodomain of the chromatin remodeling complex NoRC.
Structure, 23, 2015
|
|
3H9R
 
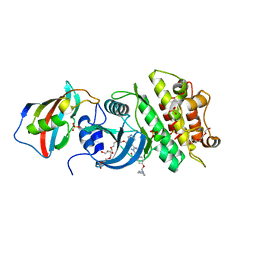 | | Crystal structure of the kinase domain of type I activin receptor (ACVR1) in complex with FKBP12 and dorsomorphin | | Descriptor: | 6-[4-(2-piperidin-1-ylethoxy)phenyl]-3-pyridin-4-ylpyrazolo[1,5-a]pyrimidine, Activin receptor type-1, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Chaikuad, A, Alfano, I, Shrestha, B, Muniz, J.R.C, Petrie, K, Fedorov, O, Phillips, C, Bishop, S, Mahajan, P, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Bone Morphogenetic Protein Receptor ALK2 and Implications for Fibrodysplasia Ossificans Progressiva.
J.Biol.Chem., 287, 2012
|
|
4HP1
 
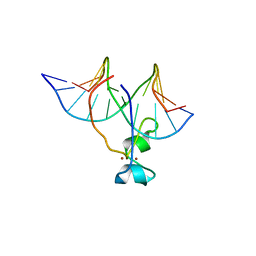 | | Crystal structure of Tet3 in complex with a non-CpG dsDNA | | Descriptor: | DNA (5'-D(*GP*CP*CP*AP*CP*(5CM)P*GP*GP*TP*GP*GP*C)-3'), LOC100036628 protein, ZINC ION | | Authors: | Chao, X, Tempel, W, Bian, C, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-10-23 | | Release date: | 2012-12-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tet3 CXXC Domain and Dioxygenase Activity Cooperatively Regulate Key Genes for Xenopus Eye and Neural Development.
Cell(Cambridge,Mass.), 151, 2012
|
|
6R7Z
 
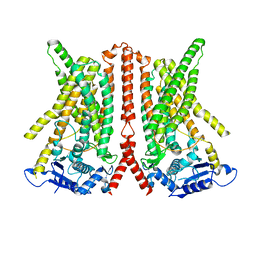 | | CryoEM structure of calcium-free human TMEM16K / Anoctamin 10 in detergent (closed form) | | Descriptor: | Anoctamin-10 | | Authors: | Pike, A.C.W, Bushell, S.R, Shintre, C.A, Tessitore, A, Chu, A, Mukhopadhyay, S, Shrestha, L, Chalk, R, Burgess-Brown, N.A, Love, J, Huiskonen, J.T, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-29 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.14 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
4QL1
 
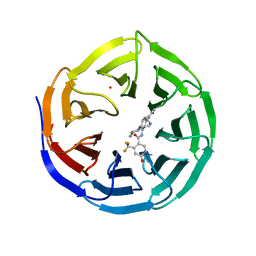 | | Crystal structure of human WDR5 in complex with compound OICR-9429 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-(4-(4-methylpiperazin-1-yl)-3'-(morpholinomethyl)-[1,1'-biphenyl]-3-yl)-6-oxo-4-(trifluoromethyl)-1,6-dihydropyridine-3-carboxamide, ... | | Authors: | Dong, A, Dombrovski, L, Walker, J.R, Getlik, M, Kuznetsova, E, Smil, D, Barsyte, D, Li, F, Poda, G, Senisterra, G, Marcellus, R, Al-Awar, R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Schapira, M, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-10 | | Release date: | 2014-12-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Pharmacological targeting of the Wdr5-MLL interaction in C/EBP alpha N-terminal leukemia.
Nat.Chem.Biol., 11, 2015
|
|
4NRB
 
 | | Crystal Structure of the bromodomain of human BAZ2B in complex with compound-1 N01197 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, N-methyl-2-(tetrahydro-2H-pyran-4-yloxy)benzamide | | Authors: | Muniz, J.R.C, Felletar, I, Chaikuad, A, Filippakopoulos, P, Ferguson, F.M, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Ciulli, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-11-26 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Targeting low-druggability bromodomains: fragment based screening and inhibitor design against the BAZ2B bromodomain.
J.Med.Chem., 56, 2013
|
|
6CCG
 
 | | Crystal structure MBD3 MBD domain in complex with methylated CpG DNA | | Descriptor: | DNA, Methyl-CpG-binding domain protein 3, UNKNOWN ATOM OR ION | | Authors: | Liu, K, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-07 | | Release date: | 2018-05-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analyses reveal that MBD3 is a methylated CG binder.
Febs J., 286, 2019
|
|
5PO7
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N11083a | | Descriptor: | 1,2-ETHANEDIOL, 1,3-dimethyl-5-(methylamino)-6-nitro-1,3-dihydro-2H-benzimidazol-2-one, Bromodomain-containing protein 1, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4IKP
 
 | | Crystal structure of coactivator-associated arginine methyltransferase 1 with methylenesinefungin | | Descriptor: | (2S,5S)-2,6-diamino-5-{[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}hexanoic acid, GLYCEROL, Histone-arginine methyltransferase CARM1, ... | | Authors: | Dong, A, Dombrovski, L, He, H, Ibanez, G, Wernimont, A, Zheng, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Luo, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-12-27 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A chemical probe of CARM1 alters epigenetic plasticity against breast cancer cell invasion.
Elife, 8, 2019
|
|
6S1F
 
 | | Structure of the kinase domain of human RIPK2 in complex with the inhibitor CSLP3 | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 2, ~{N}-[3-[2-azanyl-5-(4-piperazin-1-ylphenyl)pyridin-3-yl]-5-methoxy-phenyl]methanesulfonamide | | Authors: | Pinkas, D.M, Bufton, J.C, Kupinska, K, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-06-18 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Design of 3,5-diaryl-2-aminopyridines as receptor-interacting protein kinase 2 (RIPK2) and nucleotide-binding oligomerization domain (NOD) cell signaling inhibitors
To be published
|
|
4O45
 
 | | WDR5 in complex with influenza NS1 C-terminal tail | | Descriptor: | Nonstructural protein 1, UNKNOWN ATOM OR ION, WD repeat-containing protein 5 | | Authors: | Qin, S, Xu, C, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis for histone mimicry and hijacking of host proteins by influenza virus protein NS1.
Nat Commun, 5, 2014
|
|
3SX0
 
 | | Crystal structure of Dot1l in complex with a brominated SAH analog | | Descriptor: | (2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(4-amino-5-bromo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}sulfanyl)butanoic acid (non-preferred name), Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yu, W, Tempel, W, Smil, D, Schapira, M, Li, Y, Vedadi, M, Nguyen, K.T, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-07-14 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Bromo-deaza-SAH: a potent and selective DOT1L inhibitor.
Bioorg. Med. Chem., 21, 2013
|
|
5PO9
 
 | | PanDDA analysis group deposition -- Crystal Structure of BRD1 in complex with N07950b | | Descriptor: | 1,2-ETHANEDIOL, 1-methyl-4-phenyl-3-(trifluoromethyl)-1H-pyrazol-5-amine, Bromodomain-containing protein 1, ... | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-07 | | Release date: | 2017-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.116 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5NNG
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated SRPK1 peptide (K585ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, VAG(ALY)YS(ALY)EFFY | | Authors: | Filippakopoulos, P, Picaud, S, Newman, J, Sorrell, F, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-04-08 | | Release date: | 2018-05-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
