5R66
 
 | | PanDDA analysis group deposition -- Crystal Structure of HUMAN CLEAVAGE FACTOR IM in complex with FMOPL000198a | | Descriptor: | ACETATE ION, Cleavage and polyadenylation specificity factor subunit 5, N-(2,3-dimethylphenyl)-2-(morpholin-4-yl)acetamide, ... | | Authors: | Talon, R, Krojer, T, Diaz-Saez, L, Bradley, A.R, Aimon, A, Fairhead, M, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Huber, K.V.M, von Delft, F. | | Deposit date: | 2020-02-28 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5RZB
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z271004858 | | Descriptor: | 1,2-ETHANEDIOL, 4-amino-N-(pyridin-2-yl)benzenesulfonamide, DIMETHYL SULFOXIDE, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5RZR
 
 | | EPB41L3 PanDDA analysis group deposition -- Crystal Structure of the FERM domain of human EPB41L3 in complex with Z53825177 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, Isoform 2 of Band 4.1-like protein 3, ... | | Authors: | Bradshaw, W.J, Katis, V.L, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Gileadi, O. | | Deposit date: | 2020-10-30 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | EPB41L3 PanDDA analysis group deposition
To Be Published
|
|
5R4F
 
 | | PanDDA analysis group deposition of ground-state model of ATAD2 | | Descriptor: | 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, SULFATE ION | | Authors: | Snee, M, Talon, R, Fowley, D, Collins, P, Nelson, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Von-Delft, F. | | Deposit date: | 2020-02-21 | | Release date: | 2020-05-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | PanDDA analysis group deposition of ground-state model
To Be Published
|
|
5R59
 
 | | PanDDA analysis group deposition -- Crystal Structure of human NUDT22 in complex with N13441a | | Descriptor: | DIMETHYL SULFOXIDE, Uridine diphosphate glucose pyrophosphatase NUDT22, methyl 3-(methylsulfonylamino)benzoate | | Authors: | Diaz-Saez, L, Talon, R, Krojer, T, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K.V.M. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5R5P
 
 | | PanDDA analysis group deposition -- Crystal Structure of human NUDT22 in complex with N13910a | | Descriptor: | DIMETHYL SULFOXIDE, Uridine diphosphate glucose pyrophosphatase NUDT22, ~{N}-[(4-fluorophenyl)methyl]-4-methoxy-aniline | | Authors: | Diaz-Saez, L, Talon, R, Krojer, T, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Huber, K.V.M. | | Deposit date: | 2020-02-28 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
5R69
 
 | | XChem group deposition -- Crystal Structure of human YEATS4 in complex with XS038644e | | Descriptor: | 1,2-ETHANEDIOL, YEATS domain-containing protein 4, ~{N}-(5-oxidanylidene-7,8-dihydro-6~{H}-naphthalen-2-yl)ethanamide | | Authors: | Raux, B, Krojer, T, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Huber, K.V.M. | | Deposit date: | 2020-02-29 | | Release date: | 2020-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S8Q
 
 | | XChem group deposition -- Crystal Structure of the second bromodomain of pleckstrin homology domain interacting protein (PHIP) in complex with FMO3D000185a (space group P212121) | | Descriptor: | (3R)-3-methyl-1,2,3,4-tetrahydro-5H-1,4-benzodiazepin-5-one, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S8P
 
 | | XChem group deposition -- Crystal Structure of the second bromodomain of pleckstrin homology domain interacting protein (PHIP) in complex with E07179c (space group P212121) | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-3,4-dihydroquinazolin-2(1H)-one, CALCIUM ION, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S8O
 
 | | XChem group deposition -- Crystal Structure of the second bromodomain of pleckstrin homology domain interacting protein (PHIP) in complex with N01460c (space group P212121) | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, N-(4-chloro-2-methylphenyl)acetamide, ... | | Authors: | Krojer, T, Talon, R, Fairhead, M, Szykowska, A, Burgess-Brown, N.A, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2020-12-17 | | Release date: | 2021-01-13 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | XChem group deposition
To Be Published
|
|
3CZU
 
 | | Crystal structure of the human ephrin A2- ephrin A1 complex | | Descriptor: | Ephrin type-A receptor 2, Ephrin-A1, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Walker, J.R, Yermekbayeva, L, Seitova, A, Butler-Cole, C, Bountra, C, Wikstrom, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-30 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Architecture of Eph receptor clusters.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4LEC
 
 | | Human Methyltransferase-Like Protein 21A | | Descriptor: | Protein-lysine methyltransferase METTL21A, S-ADENOSYL-L-HOMOCYSTEINE, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Zeng, H, Fenner, M, Wernimont, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-06-25 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | The Crystal Structure of Human Methyltransferase-Like Protein 21A in Complex with SAH
To be Published
|
|
4DO0
 
 | | Crystal Structure of human PHF8 in complex with Daminozide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DAMINOZIDE, ... | | Authors: | Krojer, T, Daniel, M, Ng, S.S, Walport, L.J, Chowdhury, R, Arrowsmith, C.H, Edwards, A, Bountra, C, Kawamura, A, Muller-Knapp, S, McDonough, M.A, von Delft, F, Schofield, C.J, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: |
|
|
5O5E
 
 | | Crystal structure of human UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosaminephosphotransferase (DPAGT1) (V264G mutant) in complex with tunicamycin | | Descriptor: | (2S)-3-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-2-[(6E)-HEXADEC-6-ENOYLOXY]PROPYL (8E)-OCTADEC-8-ENOATE, Tunicamycin, UDP-N-acetylglucosamine--dolichyl-phosphate N-acetylglucosaminephosphotransferase, ... | | Authors: | Pike, A.C.W, Dong, Y.Y, Chu, A, Tessitore, A, Goubin, S, Dong, L, Mukhopadhyay, S, Mahajan, P, Chalk, R, Berridge, G, Wang, D, Kupinska, K, Belaya, K, Beeson, D, Burgess-Brown, N, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-06-01 | | Release date: | 2018-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design.
Cell, 175, 2018
|
|
4ABK
 
 | | HUMAN PARP14 (ARTD8, BAL2) - MACRO DOMAIN 3 IN COMPLEX WITH ADENOSINE- 5-DIPHOSPHORIBOSE | | Descriptor: | POLY [ADP-RIBOSE] POLYMERASE 14, [(2R,3S,4R,5R)-5-(6-AMINOPURIN-9-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHYL [HYDROXY-[[(2R,3S,4R,5S)-3,4,5-TRIHYDROXYOXOLAN-2-YL]METHOXY]PHOSPHORYL] HYDROGEN PHOSPHATE | | Authors: | Karlberg, T, Moche, M, Thorsell, A.G, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Ekblad, T, Weigelt, J, Schuler, H. | | Deposit date: | 2011-12-08 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of Mono-Adp-Ribosylated Artd10 Substrates by Artd8 Macrodomains
Structure, 21, 2013
|
|
4ABL
 
 | | HUMAN PARP14 (ARTD8, BAL2) - MACRO DOMAIN 3 | | Descriptor: | BROMIDE ION, POLY [ADP-RIBOSE] POLYMERASE 14 | | Authors: | Karlberg, T, Moche, M, Thorsell, A.G, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Ekblad, T, Weigelt, J, Schuler, H. | | Deposit date: | 2011-12-08 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Recognition of Mono-Adp-Ribosylated Artd10 Substrates by Artd8 Macrodomains
Structure, 21, 2013
|
|
4GPJ
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a isoxazolylbenzimidazole ligand | | Descriptor: | (1R)-6-(3,5-dimethyl-1,2-oxazol-4-yl)-1-phenyl-2,3-dihydro-1H-inden-1-ol, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Felletar, I, Heightman, T.D, Brennan, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The design and synthesis of 5- and 6-isoxazolylbenzimidazoles as selective inhibitors of the BET bromodomains.
Medchemcomm, 4, 2013
|
|
4C9W
 
 | | Crystal structure of NUDT1 (MTH1) with R-crizotinib | | Descriptor: | 3-[(1R)-1-(2,6-dichloro-3-fluorophenyl)ethoxy]-5-(1-piperidin-4-yl-1H-pyrazol-4-yl)pyridin-2-amine, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, CHLORIDE ION, ... | | Authors: | Elkins, J.M, Salah, E, Huber, K, Superti-Furga, G, Abdul Azeez, K.R, Raynor, J, Krojer, T, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-10-03 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stereospecific Targeting of Mth1 by (S)-Crizotinib as an Anticancer Strategy.
Nature, 508, 2014
|
|
3C8X
 
 | | Crystal structure of the ligand binding domain of human Ephrin A2 (Epha2) receptor protein kinase | | Descriptor: | Ephrin type-A receptor 2 | | Authors: | Walker, J.R, Yermekbayeva, L, Seitova, A, Butler-Cole, C, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-14 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Architecture of Eph receptor clusters.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3ZON
 
 | | Human TYK2 pseudokinase domain bound to a kinase inhibitor | | Descriptor: | 5-PHENYL-2-UREIDOTHIOPHENE-3-CARBOXAMIDE, NON-RECEPTOR TYROSINE-PROTEIN KINASE TYK2 | | Authors: | Elkins, J.M, Wang, J, Krojer, T, Savitsky, P, Chalk, R, Daga, N, Salah, E, Berridge, G, Picaud, S, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-02-22 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Human Tyk2 Pseudokinase Domain Bound to a Kinase Inhibitor
To be Published
|
|
4H12
 
 | | The crystal structure of methyltransferase domain of human SET domain-containing protein 2 in complex with S-adenosyl-L-homocysteine | | Descriptor: | CHLORIDE ION, Histone-lysine N-methyltransferase SETD2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Amaya, M.F, Dong, A, Zeng, H, Mackenzie, F, Bunnage, M, Weigelt, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-10 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Sinefungin Derivatives as Inhibitors and Structure Probes of Protein Lysine Methyltransferase SETD2.
J.Am.Chem.Soc., 134, 2012
|
|
3CE0
 
 | | Human poly(ADP-ribose) polymerase 3, catalytic fragment in complex with an inhibitor PJ34 | | Descriptor: | N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, Poly [ADP-ribose] polymerase 3 | | Authors: | Lehtio, L, Karlberg, T, Arrowsmith, C.H, Berglund, H, Bountra, C, Busam, R, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Herman, M.D, Johansson, A, Johansson, I, Kallas, A, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Persson, C, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, van den Berg, S, Welin, M, Weigelt, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-27 | | Release date: | 2008-03-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3.
J.Med.Chem., 52, 2009
|
|
4C9X
 
 | | Crystal structure of NUDT1 (MTH1) with S-crizotinib | | Descriptor: | 3-[(1S)-1-(2,6-DICHLORO-3-FLUOROPHENYL)ETHOXY]-5-(1-PIPERIDIN-4-YLPYRAZOL-4-YL)PYRIDIN-2-AMINE, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, CHLORIDE ION, ... | | Authors: | Elkins, J.M, Salah, E, Huber, K, Superti-Furga, G, Abdul Azeez, K.R, Krojer, T, von Delft, F, Bountra, C, Edwards, A, Knapp, S. | | Deposit date: | 2013-10-03 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Stereospecific Targeting of Mth1 by (S)-Crizotinib as an Anticancer Strategy.
Nature, 508, 2014
|
|
4HC4
 
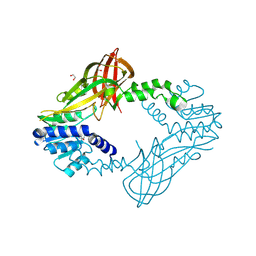 | | Human HMT1 hnRNP methyltransferase-like protein 6 (S. cerevisiae) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Protein arginine N-methyltransferase 6, ... | | Authors: | Dong, A, Zeng, H, He, H, El Bakkouri, M, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-09-28 | | Release date: | 2012-10-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis of arginine asymmetrical dimethylation by PRMT6.
Biochem. J., 473, 2016
|
|
4FMU
 
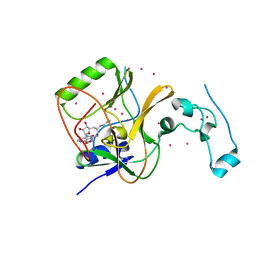 | | Crystal structure of Methyltransferase domain of human SET domain-containing protein 2 Compound: Pr-SNF | | Descriptor: | (2S,5S)-2-amino-6-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-5-(propylamino)hexanoic acid, Histone-lysine N-methyltransferase SETD2, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Zeng, H, Ibanez, G, Zheng, W, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Min, J, Luo, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-18 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Sinefungin Derivatives as Inhibitors and Structure Probes of Protein Lysine Methyltransferase SETD2.
J.Am.Chem.Soc., 134, 2012
|
|
