4RZQ
 
 | | Structural Analysis of Substrate, Reaction Intermediate and Product Binding in Haemophilus influenzae Biotin Carboxylase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Biotin carboxylase, methyl (3aS,4S,6aR)-4-(5-methoxy-5-oxopentyl)-2-oxohexahydro-1H-thieno[3,4-d]imidazole-1-carboxylate | | Authors: | Broussard, T.C, Pakhomova, S, Neau, D.B, Champion, T.S, Bonnot, R, Waldrop, G.L. | | Deposit date: | 2014-12-23 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
Biochemistry, 54, 2015
|
|
4X9Q
 
 | | MnSOD-3 Room Temperature Structure | | Descriptor: | MALONATE ION, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Hunter, G.J, Trinh, C.H, Hunter, T, Bonetta, R, Stewart, E.E. | | Deposit date: | 2014-12-11 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The structure of the Caenorhabditis elegans manganese superoxide dismutase MnSOD-3-azide complex.
Protein Sci., 24, 2015
|
|
4RDV
 
 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-FORMIMINO-L-GLUTAMATE IMINOHYDROLASE, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2014-09-19 | | Release date: | 2014-10-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate
To be Published
|
|
5UG3
 
 | | NMR SOLUTION STRUCTURE OF ALPHA-CONOTOXIN GID MUTANT A10V | | Descriptor: | Alpha-conotoxin GID | | Authors: | Hussein, A.K, Leffler, A.E, Zebroski, H.A, Powell, S.R, Kuryatov, A, Filipenko, P, Gorson, J, Heizmann, A, Lyskov, S, Nicke, A, Lindstrom, J, Rudy, B, Bonneau, R, Holford, M, Poget, S.F. | | Deposit date: | 2017-01-06 | | Release date: | 2017-09-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Discovery of peptide ligands through docking and virtual screening at nicotinic acetylcholine receptor homology models.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5UG5
 
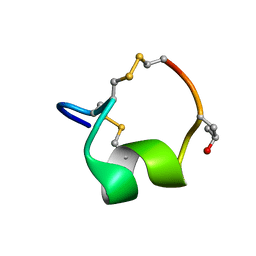 | | NMR SOLUTION STRUCTURE OF THE ALPHA-CONOTOXIN GID MUTANT V13Y | | Descriptor: | Alpha-conotoxin GID | | Authors: | Hussein, A, Leffler, A.E, Kuryatov, A, Zebroski, H.A, Powell, S.R, Filipenko, P, Gorson, J, Heizmann, A, Lyskov, S, Nicke, A, Lindstrom, J, Rudy, B, Bonneau, R, Holford, M, Poget, S.F. | | Deposit date: | 2017-01-06 | | Release date: | 2017-09-06 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Discovery of peptide ligands through docking and virtual screening at nicotinic acetylcholine receptor homology models.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7LPR
 
 | |
3MDW
 
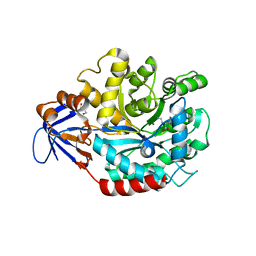 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-formimino-L-Aspartate | | Descriptor: | GLYCEROL, N-[(E)-iminomethyl]-L-aspartic acid, N-formimino-L-Glutamate Iminohydrolase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-03-30 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8979 Å) | | Cite: | Structure of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa.
Biochemistry, 54, 2015
|
|
3MDU
 
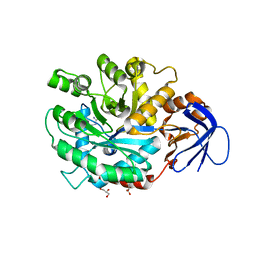 | | The structure of N-formimino-L-Glutamate Iminohydrolase from Pseudomonas aeruginosa complexed with N-Guanidino-L-Glutamate | | Descriptor: | GLYCEROL, N-carbamimidoyl-L-glutamic acid, N-formimino-L-Glutamate Iminohydrolase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Marti-Arbona, R, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-03-30 | | Release date: | 2011-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4003 Å) | | Cite: | Structure of N-Formimino-l-glutamate Iminohydrolase from Pseudomonas aeruginosa.
Biochemistry, 54, 2015
|
|
4YIN
 
 | | Crystal structure of the extended-spectrum beta-lactamase OXA-145 | | Descriptor: | Beta-lactamase, CITRATE ANION | | Authors: | Meziane-Cherif, D, Bonnet, R, Haouz, A, Courvalin, P. | | Deposit date: | 2015-03-02 | | Release date: | 2016-02-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the loss of penicillinase and the gain of ceftazidimase activities by OXA-145 beta-lactamase in Pseudomonas aeruginosa.
J. Antimicrob. Chemother., 71, 2016
|
|
4GDN
 
 | | Structure of FmtA-like protein | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Protein flp | | Authors: | Cougnoux, A, Gibold, L, Delmas, J, Robin, F, Dalmasso, G, Bonnet, R. | | Deposit date: | 2012-08-01 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Analysis of Structure-Function Relationships in the Colibactin-Maturating Enzyme ClbP.
J.Mol.Biol., 424, 2012
|
|
6DGU
 
 | | PER-2 class A extended-spectrum beta-lactamase crystal structure at 2.69 Angstrom resolution | | Descriptor: | Beta-lactamase | | Authors: | Power, P, Ruggiero, M, Gutkind, G, Bonomo, R, Klinke, S. | | Deposit date: | 2018-05-18 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.691 Å) | | Cite: | Structural Insights into the Inhibition of the Extended-Spectrum beta-Lactamase PER-2 by Avibactam.
Antimicrob.Agents Chemother., 63, 2019
|
|
4E6W
 
 | | ClbP in complex with 3-aminophenyl boronic acid | | Descriptor: | ClbP peptidase, M-AMINOPHENYLBORONIC ACID, PHOSPHATE ION | | Authors: | Cougnoux, A, Delmas, J, Bonnet, R. | | Deposit date: | 2012-03-16 | | Release date: | 2013-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The NRP peptidase ClbP as a target for the inhibition
of genotoxicity, cell proliferation and tumorogenesis
mediated by pks-harboring bacteria
To be Published
|
|
5LPR
 
 | |
5AG2
 
 | | SOD-3 azide complex | | Descriptor: | ACETATE ION, AZIDE ION, MALONATE ION, ... | | Authors: | Hunter, G.J, Trinh, C.H, Bonetta, R, Stewart, E.E, Cabelli, D.E, Hunter, T. | | Deposit date: | 2015-01-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | The Structure of the Caenorhabditis Elegans Manganese Superoxide Dismutase Mnsod-3-Azide Complex.
Protein Sci., 24, 2015
|
|
6C6I
 
 | | Crystal structure of a chimeric NDM-1 metallo-beta-lactamase harboring the IMP-1 L3 loop | | Descriptor: | Metallo-beta-lactamase type 2 chimera, ZINC ION | | Authors: | Otero, L, Giannini, E, Klinke, S, Palacios, A, Mojica, M, Bonomo, R, Llarrull, L, Vila, A. | | Deposit date: | 2018-01-18 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Reaction Mechanism of Metallo-beta-Lactamases Is Tuned by the Conformation of an Active-Site Mobile Loop.
Antimicrob. Agents Chemother., 63, 2019
|
|
6CAC
 
 | | Crystal structure of NDM-1 metallo-beta-lactamase harboring an insertion of a Pro residue in L3 loop | | Descriptor: | CADMIUM ION, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Alzari, P.M, Giannini, E, Palacios, A, Mojica, M, Bonomo, R, Llarrull, L, Vila, A. | | Deposit date: | 2018-01-30 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Reaction Mechanism of Metallo-beta-Lactamases Is Tuned by the Conformation of an Active-Site Mobile Loop.
Antimicrob. Agents Chemother., 63, 2019
|
|
6D3G
 
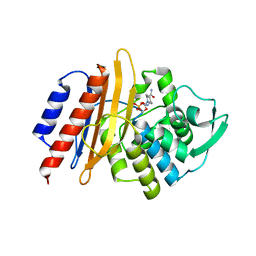 | | PER-2 class A extended-spectrum beta-lactamase crystal structure in complex with avibactam at 2.4 Angstrom resolution | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase, TETRAETHYLENE GLYCOL | | Authors: | Power, P, Ruggiero, M, Gutkind, G, Bonomo, R, Klinke, S. | | Deposit date: | 2018-04-16 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structural Insights into the Inhibition of the Extended-Spectrum beta-Lactamase PER-2 by Avibactam.
Antimicrob.Agents Chemother., 63, 2019
|
|
2AQV
 
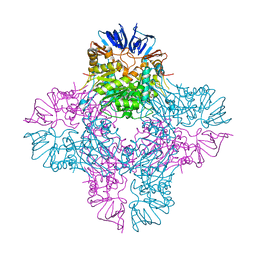 | | Crystal Structure of E. coli Isoaspartyl Dipeptidase mutant Y137F | | Descriptor: | Isoaspartyl dipeptidase, ZINC ION | | Authors: | Marti-Arbona, R, Thoden, J.B, Holden, H.M, Raushel, F.M. | | Deposit date: | 2005-08-18 | | Release date: | 2005-12-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Functional significance of Glu-77 and Tyr-137 within the active site of isoaspartyl dipeptidase.
Bioorg.Chem., 33, 2005
|
|
4E6X
 
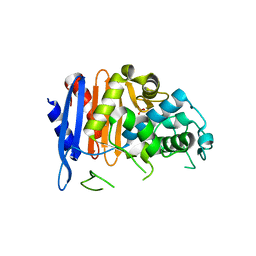 | | ClbP in complex boron-based inhibitor | | Descriptor: | ({[(chloromethyl)sulfonyl]amino}methyl)boronic acid, ClbP peptidase, PHOSPHATE ION | | Authors: | Cougnoux, A, Delmas, J, Bonnet, R. | | Deposit date: | 2012-03-16 | | Release date: | 2013-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The NRP peptidase ClbP as a target for the inhibition
of genotoxicity, cell proliferation and tumorogenesis
mediated by pks-harboring bacteria
To be Published
|
|
2PLM
 
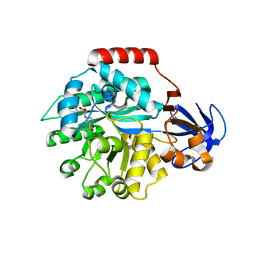 | | Crystal structure of the protein TM0936 from Thermotoga maritima complexed with ZN and S-inosylhomocysteine | | Descriptor: | (2S)-2-AMINO-4-({[(2S,3S,4R,5R)-3,4-DIHYDROXY-5-(6-OXO-1,6-DIHYDRO-9H-PURIN-9-YL)TETRAHYDROFURAN-2-YL]METHYL}THIO)BUTANOIC ACID, Uncharacterized protein, ZINC ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Hermann, J.C, Marti-Arbona, R, Shoichet, B.K, Raushel, F.M, Almo, S.C. | | Deposit date: | 2007-04-20 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based activity prediction for an enzyme of unknown function
Nature, 448, 2007
|
|
2BO9
 
 | | Human carboxypeptidase A4 in complex with human latexin. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETONE, ... | | Authors: | Pallares, I, Bonet, R, Garcia-Castellanos, R, Ventura, S, Aviles, F.X, Vendrell, J, Gomis-Rueth, F.X. | | Deposit date: | 2005-04-08 | | Release date: | 2005-04-15 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Human Carboxypeptidase A4 with its Endogenous Protein Inhibitor, Latexin.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
3HUO
 
 | | X-ray crystallographic structure of CTX-M-9 S70G in complex with benzylpenicillin | | Descriptor: | (2R,4S)-2-{(R)-carboxy[(phenylacetyl)amino]methyl}-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CTX-M-9 extended-spectrum beta-lactamase, PENICILLIN G | | Authors: | Delmas, J, Leyssene, D, Dubois, D, Robin, F, Bonnet, R. | | Deposit date: | 2009-06-15 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Dynamic view of the early and late steps of the catalytic mechanism mediated by the emerging enzymes CTX-M.
To be Published
|
|
3HRE
 
 | | X-ray crystallographic structure of CTX-M-9 S70G | | Descriptor: | CTX-M-9 extended-spectrum beta-lactamase, PHOSPHATE ION | | Authors: | Delmas, J, Leyssene, D, Dubois, D, Vazeille, E, Robin, F, Bonnet, R. | | Deposit date: | 2009-06-09 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into substrate recognition and product expulsion in CTX-M enzymes.
J.Mol.Biol., 400, 2010
|
|
3HLW
 
 | | CTX-M-9 S70G in complex with cefotaxime | | Descriptor: | (6R,7R)-3-(acetyloxymethyl)-7-[[(2Z)-2-(2-amino-1,3-thiazol-4-yl)-2-methoxyimino-ethanoyl]amino]-8-oxo-5-thia-1-azabicy clo[4.2.0]oct-2-ene-2-carboxylic acid, CTX-M-9 extended-spectrum beta-lactamase | | Authors: | Delmas, J, Leyssne, D, Dubois, D, Vazeille, E, Robin, F, Bonnet, R. | | Deposit date: | 2009-05-28 | | Release date: | 2010-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into substrate recognition and product expulsion in CTX-M enzymes.
J.Mol.Biol., 400, 2010
|
|
3HVF
 
 | | X-ray crystallographic structure of CTX-M-9 S70G in complex with hydrolyzed benzylpenicillin | | Descriptor: | (2R,4S)-2-{(R)-carboxy[(phenylacetyl)amino]methyl}-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CTX-M-9 extended-spectrum beta-lactamase | | Authors: | Delmas, J, Leyssene, D, Dubois, D, Vazeille, E, Robin, F, Bonnet, R. | | Deposit date: | 2009-06-16 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into substrate recognition and product expulsion in CTX-M enzymes.
J.Mol.Biol., 400, 2010
|
|
