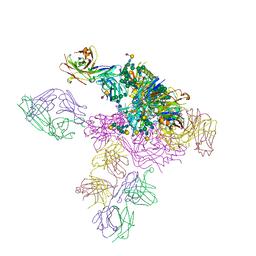
5D4K
 
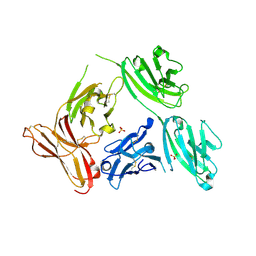 | |
5F1S
 
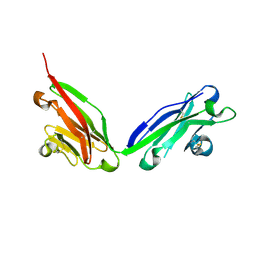 | |
5F7E
 
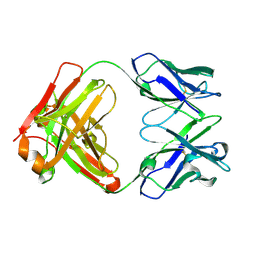 | | Crystal structure of germ-line precursor of 3BNC60 Fab | | Descriptor: | Fab heavy chain, Fab light chain | | Authors: | Sievers, S.A, Scharf, L, Jiang, S, Bjorkman, P.J. | | Deposit date: | 2015-12-08 | | Release date: | 2016-04-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for germline antibody recognition of HIV-1 immunogens.
Elife, 5, 2016
|
|
5T3X
 
 | |
5UEL
 
 | | Crystal structure of 354NC102 Fab | | Descriptor: | 354NC102 Fab Heavy Chain, 354NC102 Fab Light Chain, SULFATE ION | | Authors: | Sievers, S.A, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2017-01-02 | | Release date: | 2017-02-01 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller.
Sci Transl Med, 9, 2017
|
|
5UD9
 
 | | Crystal structure of 354BG18 Fab | | Descriptor: | Fab heavy chain, Light chain | | Authors: | Scharf, L, Bjorkman, P.J. | | Deposit date: | 2016-12-23 | | Release date: | 2017-02-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller.
Sci Transl Med, 9, 2017
|
|
5VIY
 
 | |
5VIC
 
 | | Crystal structure of anti-Zika antibody Z004 bound to DENV-1 Envelope protein DIII | | Descriptor: | Dengue 1 Envelope DIII domain, Fab heavy chain, Fab light chain | | Authors: | Keeffe, J.R, West Jr, A.P, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2017-04-14 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recurrent Potent Human Neutralizing Antibodies to Zika Virus in Brazil and Mexico.
Cell, 169, 2017
|
|
5THR
 
 | | Cryo-EM structure of a BG505 Env-sCD4-17b-8ANC195 complex | | Descriptor: | 17b Fab VH domain, 17b Fab VL domain, 8ANC195 G52K5 VH domain, ... | | Authors: | Wang, H, Bjorkman, P.J. | | Deposit date: | 2016-09-30 | | Release date: | 2016-11-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Cryo-EM structure of a CD4-bound open HIV-1 envelope trimer reveals structural rearrangements of the gp120 V1V2 loop.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5VIG
 
 | | Crystal structure of anti-Zika antibody Z006 bound to Zika virus envelope protein DIII | | Descriptor: | CITRATE ANION, Fab heavy chain, Fab light chain, ... | | Authors: | Keeffe, J.R, West Jr, A.P, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2017-04-16 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recurrent Potent Human Neutralizing Antibodies to Zika Virus in Brazil and Mexico.
Cell, 169, 2017
|
|
5VJ6
 
 | |
5UEM
 
 | | Crystal structure of 354NC37 Fab in complex with HIV-1 clade AE strain 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 354NC37 Fab Heavy Chain, ... | | Authors: | Sievers, S.A, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2017-01-02 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Coexistence of potent HIV-1 broadly neutralizing antibodies and antibody-sensitive viruses in a viremic controller.
Sci Transl Med, 9, 2017
|
|
5T3Z
 
 | |
5VVF
 
 | | Crystal Structure of 354BG1 Fab | | Descriptor: | 1,2-ETHANEDIOL, 354BG1 Heavy Chain, 354BG1 Light Chain | | Authors: | Scharf, L, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2017-05-19 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Asymmetric recognition of HIV-1 Envelope trimer by V1V2 loop-targeting antibodies.
Elife, 6, 2017
|
|
7K90
 
 | | Structure of the SARS-CoV-2 S 6P trimer in complex with the human neutralizing antibody Fab fragment, C144 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C144 Fab Heavy Chain, C144 Fab Light Chain, ... | | Authors: | Barnes, C.O, Esswein, S.R, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
1DQO
 
 | | Crystal structure of the cysteine rich domain of mannose receptor complexed with Acetylgalactosamine-4-sulfate | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose, MANNOSE RECEPTOR | | Authors: | Liu, Y, Chirino, A.J, Misulovin, Z, Leteux, C, Feizi, T, Nussenzweig, M.C, Bjorkman, P.J. | | Deposit date: | 2000-01-04 | | Release date: | 2000-05-10 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the cysteine-rich domain of mannose receptor complexed with a sulfated carbohydrate ligand.
J.Exp.Med., 191, 2000
|
|
7KDE
 
 | |
7KL9
 
 | |
7K8S
 
 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C002 (state 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C002 Fab Heavy Chain, ... | | Authors: | Barnes, C.O, Malyutin, A.G, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8P
 
 | |
7K8M
 
 | | Structure of the SARS-CoV-2 receptor binding domain in complex with the human neutralizing antibody Fab fragment, C102 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C102 Fab Heavy Chain, C102 Fab Light Chain, ... | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8V
 
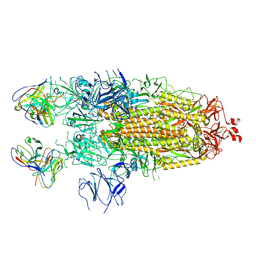 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C110 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C110 Fab Heavy Chain, ... | | Authors: | Dam, K.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8O
 
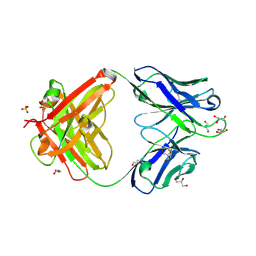 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C002 | | Descriptor: | C002 Fab Heavy Chain, C002 Fab Light Chain, GLYCEROL, ... | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
7K8W
 
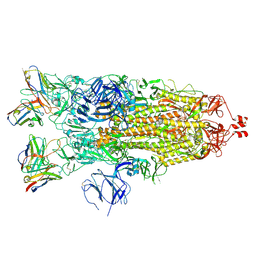 | | Structure of the SARS-CoV-2 S 2P trimer in complex with the human neutralizing antibody Fab fragment, C119 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C119 Fab Heavy Chain, ... | | Authors: | Sharaf, N.G, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
1CFB
 
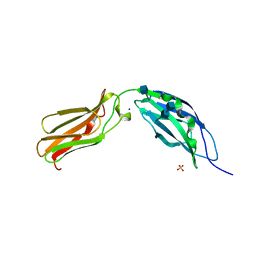 | | CRYSTAL STRUCTURE OF TANDEM TYPE III FIBRONECTIN DOMAINS FROM DROSOPHILA NEUROGLIAN AT 2.0 ANGSTROMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DROSOPHILA NEUROGLIAN, ... | | Authors: | Huber, A.H, Wang, Y.E, Bieber, A.J, Bjorkman, P.J. | | Deposit date: | 1994-08-27 | | Release date: | 1994-11-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of tandem type III fibronectin domains from Drosophila neuroglian at 2.0 A.
Neuron, 12, 1994
|
|
