1HLA
 
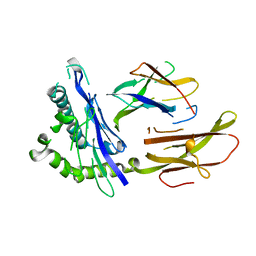 | | STRUCTURE OF THE HUMAN CLASS I HISTOCOMPATIBILITY ANTIGEN, HLA-A2 | | Descriptor: | BETA 2-MICROGLOBULIN, CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A2) (ALPHA CHAIN) | | Authors: | Bjorkman, P.J, Saper, M.A, Samraoui, B, Bennett, W.S, Strominger, J.L, Wiley, D.C. | | Deposit date: | 1987-10-15 | | Release date: | 1988-01-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the human class I histocompatibility antigen, HLA-A2.
Nature, 329, 1987
|
|
1CWV
 
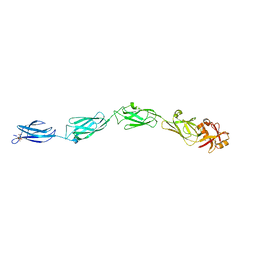 | |
1I4H
 
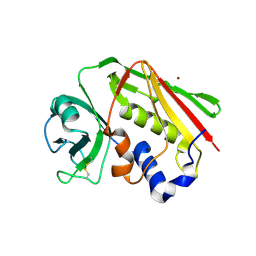 | | Crystal structure of Zn2+ soaked Staphylococcal enterotoxin A mutant H187A | | Descriptor: | ENTEROTOXIN TYPE A, ZINC ION | | Authors: | Hakansson, M, Antonsson, P, Bjork, P, Svensson, L.A. | | Deposit date: | 2001-02-21 | | Release date: | 2001-02-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Cooperative zinc binding in a staphylococcal enterotoxin A mutant mimics the SEA-MHC class II interaction
J.Biol.Inorg.Chem., 6, 2001
|
|
1I4G
 
 | | Crystal structure of Staphylococcal enterotoxin A mutant H187A with reduced Zn2+ affinity | | Descriptor: | ENTEROTOXIN TYPE A, SULFATE ION, ZINC ION | | Authors: | Hakansson, M, Antonsson, P, Bjork, P, Svensson, L.A. | | Deposit date: | 2001-02-21 | | Release date: | 2001-02-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative zinc binding in a staphylococcal enterotoxin A mutant mimics the SEA-MHC class II interaction
J.Biol.Inorg.Chem., 6, 2001
|
|
1ENF
 
 | | CRYSTAL STRUCTURE OF STAPHYLOCOCCAL ENTEROTOXIN H DETERMINED TO 1.69 A RESOLUTION | | Descriptor: | ENTEROTOXIN H, SULFATE ION | | Authors: | Hakansson, M, Petersson, K, Nilsson, H, Forsberg, G, Bjork, P, Antonsson, P, Svensson, A. | | Deposit date: | 2000-03-21 | | Release date: | 2000-04-19 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | The crystal structure of staphylococcal enterotoxin H: implications for binding properties to MHC class II and TcR molecules.
J.Mol.Biol., 302, 2000
|
|
1EWC
 
 | | CRYSTAL STRUCTURE OF ZN2+ LOADED STAPHYLOCOCCAL ENTEROTOXIN H | | Descriptor: | ENTEROTOXIN H, ZINC ION | | Authors: | Hakansson, M, Petersson, K, Nilsson, H, Forsberg, G, Bjork, P, Antonsson, P, Svensson, A. | | Deposit date: | 2000-04-25 | | Release date: | 2000-05-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of staphylococcal enterotoxin H: implications for binding properties to MHC class II and TcR molecules.
J.Mol.Biol., 302, 2000
|
|
1F77
 
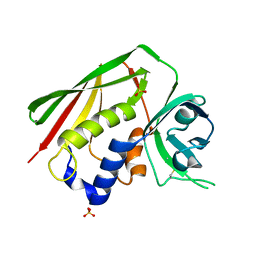 | | STAPHYLOCOCCAL ENTEROTOXIN H DETERMINED TO 2.4 A RESOLUTION | | Descriptor: | ENTEROTOXIN H, SULFATE ION | | Authors: | Hakansson, M, Petersson, K, Nilsson, H, Forsberg, G, Bjork, P. | | Deposit date: | 2000-06-26 | | Release date: | 2000-07-19 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of staphylococcal enterotoxin H: implications for binding properties to MHC class II and TcR molecules.
J.Mol.Biol., 302, 2000
|
|
7RKS
 
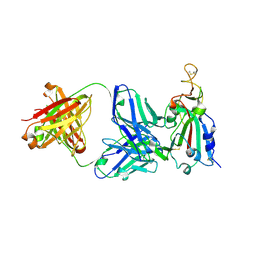 | | Structure of the SARS-CoV receptor binding domain in complex with the human neutralizing antibody Fab fragment, C118 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, C118 Antibody Fab Heavy Chain, ... | | Authors: | Jette, C.A, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
7RKV
 
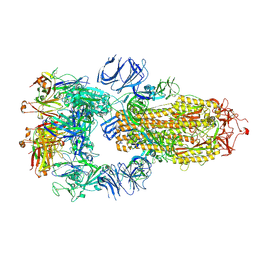 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C118 (State 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C118 Fab Heavy Chain, C118 Fab Light Chain, ... | | Authors: | Barnes, C.O, Jette, C.A, Bjorkman, P.J. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
7RKU
 
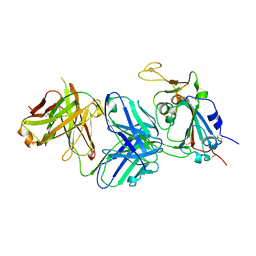 | | Structure of the SARS-CoV-2 receptor binding domain in complex with the human neutralizing antibody Fab fragment, C022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C022 Antibody Fab Heavy Chain, C022 Antibody Fab Light Chain, ... | | Authors: | Jette, C.A, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
4NHG
 
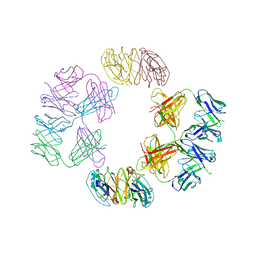 | | Crystal Structure of 2G12 IgG Dimer | | Descriptor: | 2G12 IgG dimer heavy chain, 2G12 IgG dimer light chain, Hepatitis B virus receptor binding protein | | Authors: | Wu, Y, West Jr, A.P, Kim, H.J, Thornton, M.E, Ward, A.B, Bjorkman, P.J. | | Deposit date: | 2013-11-05 | | Release date: | 2014-02-26 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (8.001 Å) | | Cite: | Structural basis for enhanced HIV-1 neutralization by a dimeric immunoglobulin G form of the glycan-recognizing antibody 2G12.
Cell Rep, 5, 2013
|
|
3FRU
 
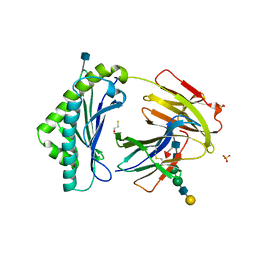 | | NEONATAL FC RECEPTOR, PH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-2-MICROGLOBULIN, BETA-MERCAPTOETHANOL, ... | | Authors: | Vaughn, D.E, Burmeister, W.P, Bjorkman, P.J. | | Deposit date: | 1997-12-22 | | Release date: | 1998-06-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of pH-dependent antibody binding by the neonatal Fc receptor.
Structure, 6, 1998
|
|
7M6F
 
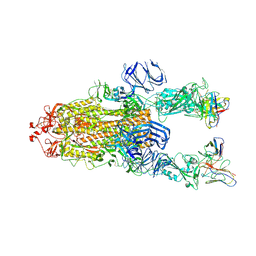 | |
7M6G
 
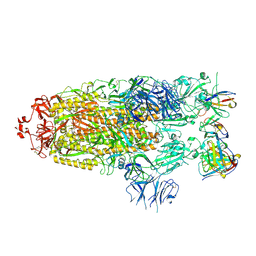 | |
7M6I
 
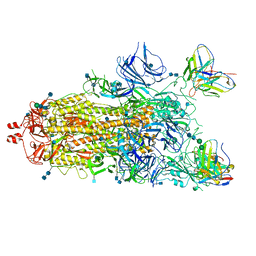 | |
7M6E
 
 | |
7M6H
 
 | |
7M6D
 
 | |
6UDJ
 
 | | HIV-1 bNAb 1-18 in complex with BG505 SOSIP.664 and 10-1074 | | Descriptor: | 1-18 Fab Heavy Chain, 1-18 Fab Light Chain, 10-1074 Fab Heavy Chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2019-09-19 | | Release date: | 2020-01-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Restriction of HIV-1 Escape by a Highly Broad and Potent Neutralizing Antibody.
Cell, 180, 2020
|
|
6UDK
 
 | | HIV-1 bNAb 1-55 in complex with modified BG505 SOSIP-based immunogen RC1 and 10-1074 | | Descriptor: | 1-55 Fab Heavy Chain, 1-55 Fab Light Chain, 10-1074 Fab Heavy Chain, ... | | Authors: | Abernathy, M.E, Barnes, C.O, Gristick, H.B, Bjorkman, P.J. | | Deposit date: | 2019-09-19 | | Release date: | 2020-01-29 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Restriction of HIV-1 Escape by a Highly Broad and Potent Neutralizing Antibody.
Cell, 180, 2020
|
|
1G0X
 
 | |
6DFJ
 
 | |
7RFB
 
 | | Crystal structure of broadly neutralizing antibody mAb1198 in complex with Hepatitis C virus envelope glycoprotein E2 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Flyak, A.I, Bjorkman, P.J. | | Deposit date: | 2021-07-14 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Analysis of antibodies from HCV elite neutralizers identifies genetic determinants of broad neutralization.
Immunity, 55, 2022
|
|
7RFC
 
 | |
1FWV
 
 | |
