7NNL
 
 | | Cryo-EM structure of the KdpFABC complex in an E1-ATP conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Silberberg, J.M, Corey, R.A, Hielkema, L, Stock, C, Stansfeld, P.J, Paulino, C, Haenelt, I. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Deciphering ion transport and ATPase coupling in the intersubunit tunnel of KdpFABC.
Nat Commun, 12, 2021
|
|
7NNP
 
 | | Rb-loaded cryo-EM structure of the E1-ATP KdpFABC complex. | | Descriptor: | CARDIOLIPIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Silberberg, J.M, Corey, R.A, Hielkema, L, Stock, C, Stansfeld, P.J, Paulino, C, Haenelt, I. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Deciphering ion transport and ATPase coupling in the intersubunit tunnel of KdpFABC.
Nat Commun, 12, 2021
|
|
6DUQ
 
 | | Structure of a Rho-NusG KOW domain complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Berger, J.M, Lawson, M.R. | | Deposit date: | 2018-06-21 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanism for the Regulated Control of Bacterial Transcription Termination by a Universal Adaptor Protein.
Mol. Cell, 71, 2018
|
|
1BGW
 
 | | TOPOISOMERASE RESIDUES 410-1202, | | Descriptor: | TOPOISOMERASE | | Authors: | Berger, J.M, Gamblin, S.J, Harrison, S.C, Wang, J.C. | | Deposit date: | 1996-02-20 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure and mechanism of DNA topoisomerase II.
Nature, 379, 1996
|
|
1MEY
 
 | | CRYSTAL STRUCTURE OF A DESIGNED ZINC FINGER PROTEIN BOUND TO DNA | | Descriptor: | CHLORIDE ION, CONSENSUS ZINC FINGER, DNA (5'-D(*AP*TP*GP*AP*GP*GP*CP*AP*GP*AP*AP*CP*T)-3'), ... | | Authors: | Kim, C.A, Berg, J.M. | | Deposit date: | 1996-09-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A 2.2 A Resolution Crystal Structure of a Designed Zinc Finger Protein Bound to DNA
Nat.Struct.Biol., 3, 1996
|
|
1FCH
 
 | | CRYSTAL STRUCTURE OF THE PTS1 COMPLEXED TO THE TPR REGION OF HUMAN PEX5 | | Descriptor: | PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR, PTS1-CONTAINING PEPTIDE | | Authors: | Gatto Jr, G.J, Geisbrecht, B.V, Gould, S.J, Berg, J.M. | | Deposit date: | 2000-07-18 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5.
Nat.Struct.Biol., 7, 2000
|
|
1M9O
 
 | |
1PXE
 
 | |
3GFP
 
 | | Structure of the C-terminal domain of the DEAD-box protein Dbp5 | | Descriptor: | DEAD box protein 5 | | Authors: | Erzberger, J.P, Dossani, Z.Y, Weirich, C.S, Weis, K, Berger, J.M. | | Deposit date: | 2009-02-27 | | Release date: | 2009-09-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the C-terminus of the mRNA export factor Dbp5 reveals the interaction surface for the ATPase activator Gle1
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4FM9
 
 | | Human topoisomerase II alpha bound to DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(P*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), DNA topoisomerase 2-alpha, ... | | Authors: | Wendorff, T.J, Schmidt, B.H, Heslop, P, Austin, C.A, Berger, J.M. | | Deposit date: | 2012-06-15 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | The Structure of DNA-Bound Human Topoisomerase II Alpha: Conformational Mechanisms for Coordinating Inter-Subunit Interactions with DNA Cleavage.
J.Mol.Biol., 424, 2012
|
|
3UC1
 
 | | Mycobacterium tuberculosis gyrase type IIA topoisomerase C-terminal domain | | Descriptor: | ACETATE ION, CALCIUM ION, DNA gyrase subunit A, ... | | Authors: | Tretter, E.M, Berger, J.M. | | Deposit date: | 2011-10-25 | | Release date: | 2012-03-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanisms for Defining Supercoiling Set Point of DNA Gyrase Orthologs: II. THE SHAPE OF THE GyrA SUBUNIT C-TERMINAL DOMAIN (CTD) IS NOT A SOLE DETERMINANT FOR CONTROLLING SUPERCOILING EFFICIENCY.
J.Biol.Chem., 287, 2012
|
|
3L4K
 
 | | Topoisomerase II-DNA cleavage complex, metal-bound | | Descriptor: | 3'-THIO-THYMIDINE-5'-PHOSPHATE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*CP*GP*TP*CP*AP*TP*CP*C)-3'), DNA (5'-D(*CP*GP*CP*GP*GP*TP*AP*GP*CP*AP*GP*TP*AP*GP*G)-3'), ... | | Authors: | Schmidt, B.H, Burgin, A.B, Deweese, J.E, Osheroff, N, Berger, J.M. | | Deposit date: | 2009-12-20 | | Release date: | 2010-05-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | A novel and unified two-metal mechanism for DNA cleavage by type II and IA topoisomerases.
Nature, 465, 2010
|
|
7ZRG
 
 | | Cryo-EM map of the WT KdpFABC complex in the E1_ATPearly conformation, under turnover conditions | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CARDIOLIPIN, POTASSIUM ION, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Rheinberger, J, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7UGW
 
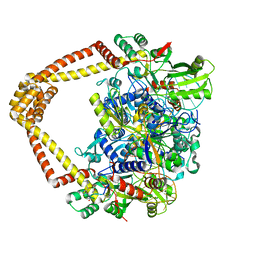 | | M. tuberculosis DNA gyrase cleavage core bound to DNA and evybactin | | Descriptor: | DNA (46-MER), DNA gyrase subunit A, DNA gyrase subunit B, ... | | Authors: | Hauk, G, Imai, Y, Lewis, K, Berger, J.M. | | Deposit date: | 2022-03-25 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-02 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Evybactin is a DNA gyrase inhibitor that selectively kills Mycobacterium tuberculosis.
Nat.Chem.Biol., 18, 2022
|
|
3C4O
 
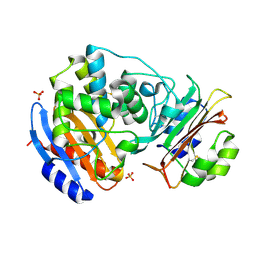 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) E73M/S130K/S146M complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein, SULFATE ION | | Authors: | Reynolds, K.A, Hanes, M.S, Thomson, J.M, Antczak, A.J, Berger, J.M, Bonomo, R.A, Kirsch, J.F, Handel, T.M. | | Deposit date: | 2008-01-30 | | Release date: | 2008-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational redesign of the SHV-1 beta-lactamase/beta-lactamase inhibitor protein interface.
J.Mol.Biol., 382, 2008
|
|
3C4P
 
 | | Crystal Structure of the SHV-1 Beta-lactamase/Beta-lactamase inhibitor protein (BLIP) E73M complex | | Descriptor: | Beta-lactamase SHV-1, Beta-lactamase inhibitory protein, SULFATE ION | | Authors: | Reynolds, K.A, Hanes, M.S, Thomson, J.M, Antczak, A.J, Berger, J.M, Bonomo, R.A, Kirsch, J.F, Handel, T.M. | | Deposit date: | 2008-01-30 | | Release date: | 2008-05-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Computational redesign of the SHV-1 beta-lactamase/beta-lactamase inhibitor protein interface.
J.Mol.Biol., 382, 2008
|
|
4TXY
 
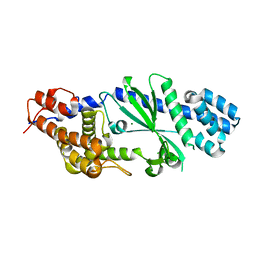 | | Crystal structure of Vibrio cholerae DncV cyclic AMP-GMP synthase, a prokaryotic cGAS homolog | | Descriptor: | Cyclic AMP-GMP synthase, MAGNESIUM ION | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Wilson, S.C, Solovykh, M.S, Vance, R.E, Berger, J.M, Doudna, J.A. | | Deposit date: | 2014-07-07 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.0001 Å) | | Cite: | Structure-Guided Reprogramming of Human cGAS Dinucleotide Linkage Specificity.
Cell, 158, 2014
|
|
4TY0
 
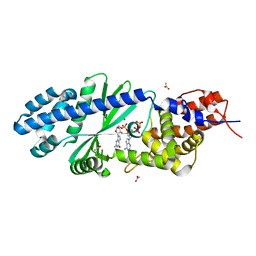 | | Crystal structure of Vibrio cholerae DncV cyclic AMP-GMP synthase in complex with linear intermediate 5' pppA(3',5')pG | | Descriptor: | ACETATE ION, Cyclic AMP-GMP synthase, MAGNESIUM ION, ... | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Wilson, S.C, Solovykh, M.S, Vance, R.E, Berger, J.M, Doudna, J.A. | | Deposit date: | 2014-07-07 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-Guided Reprogramming of Human cGAS Dinucleotide Linkage Specificity.
Cell, 158, 2014
|
|
4TXZ
 
 | | Crystal structure of Vibrio cholerae DncV cyclic AMP-GMP synthase in complex with nonhydrolyzable GTP | | Descriptor: | Cyclic AMP-GMP synthase, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Wilson, S.C, Solovykh, M.S, Vance, R.E, Berger, J.M, Doudna, J.A. | | Deposit date: | 2014-07-07 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Guided Reprogramming of Human cGAS Dinucleotide Linkage Specificity.
Cell, 158, 2014
|
|
4WMG
 
 | | Structure of hen egg-white lysozyme from a microfludic harvesting device using synchrotron radiation (2.5A) | | Descriptor: | Lysozyme C | | Authors: | Lyubimov, A.Y, Murray, T.D, Koehl, A, Uervirojnangkoorn, M, Zeldin, O.B, Cohen, A.E, Soltis, S.M, Baxter, E.M, Brewster, A.S, Sauter, N.K, Brunger, A.T, Berger, J.M. | | Deposit date: | 2014-10-08 | | Release date: | 2015-04-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Capture and X-ray diffraction studies of protein microcrystals in a microfluidic trap array.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4XGC
 
 | | Crystal structure of the eukaryotic origin recognition complex | | Descriptor: | CHLORIDE ION, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | Authors: | Bleichert, F, Botchan, M.R, Berger, J.M. | | Deposit date: | 2014-12-30 | | Release date: | 2015-04-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the eukaryotic origin recognition complex.
Nature, 519, 2015
|
|
7ZRM
 
 | | Cryo-EM map of the unphosphorylated KdpFABC complex in the E1-P_ADP conformation, under turnover conditions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRD
 
 | | Cryo-EM map of the WT KdpFABC complex in the E1-P tight conformation, stabilised with the inhibitor orthovanadate | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRL
 
 | | Cryo-EM map of the unphosphorylated KdpFABC complex in the E2-P conformation, under turnover conditions | | Descriptor: | POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, Potassium-transporting ATPase KdpC subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Dubach, V.R.A, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
7ZRH
 
 | | Cryo-EM structure of the KdpFABC complex in a nucleotide-free E1 conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, POTASSIUM ION, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Hielkema, L, Stock, C, Silberberg, J.M, Corey, R.A, Wunnicke, D, Stansfeld, P.J, Haenelt, I, Paulino, C. | | Deposit date: | 2022-05-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Inhibited KdpFABC transitions into an E1 off-cycle state.
Elife, 11, 2022
|
|
