7K4F
 
 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor 31 | | Descriptor: | 5-[(4-{cis-4-[3-(trifluoromethyl)phenyl]cyclohexyl}piperazin-1-yl)methyl]pyridin-2(1H)-one, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
7K4D
 
 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor 3OG | | Descriptor: | 5-[(4-{trans-4-hydroxy-4-[3-(trifluoromethyl)phenyl]cyclohexyl}piperazin-1-yl)methyl]pyridin-2(1H)-one, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
7K4B
 
 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor cis-22a | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-[cis-4-(3-methylphenyl)cyclohexyl]-4-(pyridin-3-yl)piperazine, CALCIUM ION, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
7K4C
 
 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor Br-cis-22a | | Descriptor: | 1-(5-bromopyridin-3-yl)-4-[cis-4-(3-methylphenyl)cyclohexyl]piperazine, CALCIUM ION, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
7K4E
 
 | | Cryo-EM structure of human TRPV6 in complex with (4- phenylcyclohexyl)piperazine inhibitor 30 | | Descriptor: | 5-({4-[(1R,4S)-3'-methyl[1,2,3,4-tetrahydro[1,1'-biphenyl]]-4-yl]piperazin-1-yl}methyl)pyridin-2(1H)-one, Transient receptor potential cation channel subfamily V member 6 | | Authors: | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.34 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
7K4A
 
 | | Cryo-EM structure of human TRPV6 in the open state | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CALCIUM ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Neuberger, A, Nadezhdin, K.D, Singh, A.K, Sobolevsky, A.I. | | Deposit date: | 2020-09-15 | | Release date: | 2020-12-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Inactivation-mimicking block of the epithelial calcium channel TRPV6.
Sci Adv, 6, 2020
|
|
5P9J
 
 | | BTK1 COCRYSTALLIZED WITH IBRUTINIB | | Descriptor: | 1-[(3~{R})-3-[4-azanyl-3-(4-phenoxyphenyl)pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl]propan-1-one, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A.S. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-24 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Ability of Bruton's Tyrosine Kinase Inhibitors to Sequester Y551 and Prevent Phosphorylation Determines Potency for Inhibition of Fc Receptor but not B-Cell Receptor Signaling.
Mol. Pharmacol., 91, 2017
|
|
5P9L
 
 | | BTK1 IN COMPLEX WITH CC 292 | | Descriptor: | Tyrosine-protein kinase BTK, ~{N}-[3-[[5-fluoranyl-2-[[4-(2-methoxyethoxy)phenyl]amino]pyrimidin-4-yl]amino]phenyl]propanamide | | Authors: | Gardberg, A.S. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-24 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Ability of Bruton's Tyrosine Kinase Inhibitors to Sequester Y551 and Prevent Phosphorylation Determines Potency for Inhibition of Fc Receptor but not B-Cell Receptor Signaling.
Mol. Pharmacol., 91, 2017
|
|
5P9F
 
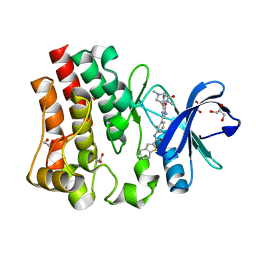 | | BTK IN COMPLEX WITH GDC-0834 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-{3-[6-({4-[(2R)-1,4-dimethyl-3-oxopiperazin-2-yl]phenyl}amino)-4-methyl-5-oxo-4,5-dihydropyrazin-2-yl]-2-methylphenyl }-4,5,6,7-tetrahydro-1-benzothiophene-2-carboxamide, ... | | Authors: | Gardberg, A.S. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Ability of Bruton's Tyrosine Kinase Inhibitors to Sequester Y551 and Prevent Phosphorylation Determines Potency for Inhibition of Fc Receptor but not B-Cell Receptor Signaling.
Mol. Pharmacol., 91, 2017
|
|
5P9G
 
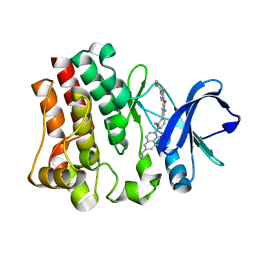 | | Structure of BTK with RN486 | | Descriptor: | 6-cyclopropyl-8-fluoranyl-2-[2-(hydroxymethyl)-3-[1-methyl-5-[[5-(4-methylpiperazin-1-yl)pyridin-2-yl]amino]-6-oxidanylidene-pyridin-3-yl]phenyl]isoquinolin-1-one, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A.S. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ability of Bruton's Tyrosine Kinase Inhibitors to Sequester Y551 and Prevent Phosphorylation Determines Potency for Inhibition of Fc Receptor but not B-Cell Receptor Signaling.
Mol. Pharmacol., 91, 2017
|
|
5P9M
 
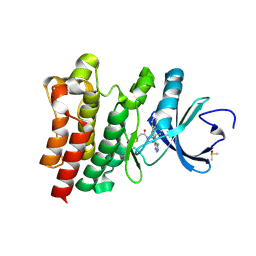 | | BTK1 BINDS COVALENTLY TO HY-15771 ONO-4059 | | Descriptor: | 6-azanyl-9-[(3~{R})-1-[(~{E})-but-2-enoyl]pyrrolidin-3-yl]-7-(4-phenoxyphenyl)purin-8-one, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A.S. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-24 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Ability of Bruton's Tyrosine Kinase Inhibitors to Sequester Y551 and Prevent Phosphorylation Determines Potency for Inhibition of Fc Receptor but not B-Cell Receptor Signaling.
Mol. Pharmacol., 91, 2017
|
|
5P9I
 
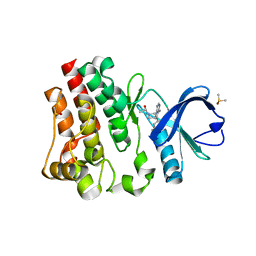 | | BTK1 SOAKED WITH IBRUTINIB-Rev | | Descriptor: | 1-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}prop-2-en-1-one, DIMETHYL SULFOXIDE, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A.S. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Ability of Bruton's Tyrosine Kinase Inhibitors to Sequester Y551 and Prevent Phosphorylation Determines Potency for Inhibition of Fc Receptor but not B-Cell Receptor Signaling.
Mol. Pharmacol., 91, 2017
|
|
5P9K
 
 | | CRYSTAL STRUCTURE OF BTK with CNX 774 | | Descriptor: | 4-[4-[[5-fluoranyl-4-[[3-(propanoylamino)phenyl]amino]pyrimidin-2-yl]amino]phenoxy]-~{N}-methyl-pyridine-2-carboxamide, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A.S. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Ability of Bruton's Tyrosine Kinase Inhibitors to Sequester Y551 and Prevent Phosphorylation Determines Potency for Inhibition of Fc Receptor but not B-Cell Receptor Signaling.
Mol. Pharmacol., 91, 2017
|
|
5P9H
 
 | | BTK1 COCRYSTALLIZED WITH RN983 | | Descriptor: | 6-~{tert}-butyl-8-fluoranyl-2-[3-(hydroxymethyl)-4-[1-methyl-5-[[5-(1-methylpiperidin-4-yl)pyridin-2-yl]amino]-6-oxidanylidene-pyridazin-3-yl]pyridin-2-yl]phthalazin-1-one, Tyrosine-protein kinase BTK | | Authors: | Gardberg, A.S. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Ability of Bruton's Tyrosine Kinase Inhibitors to Sequester Y551 and Prevent Phosphorylation Determines Potency for Inhibition of Fc Receptor but not B-Cell Receptor Signaling.
Mol. Pharmacol., 91, 2017
|
|
3TM0
 
 | | Crystal Structure of 3',5"-Aminoglycoside Phosphotransferase Type IIIa AMPPNP Butirosin A Complex | | Descriptor: | (2S)-4-amino-N-[(1R,2S,3R,4R,5S)-5-amino-4-[(2,6-diamino-2,6-dideoxy-alpha-D-glucopyranosyl)oxy]-2-hydroxy-3-(beta-D-xylofuranosyloxy)cyclohexyl]-2-hydroxybutanamide, Aminoglycoside 3'-phosphotransferase, MAGNESIUM ION, ... | | Authors: | Berghuis, A.M, Fong, D.H. | | Deposit date: | 2011-08-30 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of APH(3')-IIIa-mediated resistance to N1-substituted aminoglycoside antibiotics.
Antimicrob.Agents Chemother., 53, 2009
|
|
4DGQ
 
 | | Crystal structure of Non-heme chloroperoxidase from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, Non-heme chloroperoxidase | | Authors: | Gardberg, A.S, Edwards, T.E, Abendroth, J.A, Staker, B, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-01-26 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Non-heme chloroperoxidase from Burkholderia cenocepacia
To be Published
|
|
4HW4
 
 | |
3Q2M
 
 | | Crystal Structure of Spectinomycin Phosphotransferase, APH(9)-Ia, Protein Kinase Inhibitor CKI-7 Complex | | Descriptor: | N-(2-AMINOETHYL)-5-CHLOROISOQUINOLINE-8-SULFONAMIDE, NICKEL (II) ION, Spectinomycin phosphotransferase | | Authors: | Berghuis, A.M, Fong, D.H, Xiong, B, Hwang, J. | | Deposit date: | 2010-12-20 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of two aminoglycoside kinases bound with a eukaryotic protein kinase inhibitor.
Plos One, 6, 2011
|
|
8A1E
 
 | | Rabies virus glycoprotein in complex with Fab fragments of 17C7 and 1112-1 neutralizing antibodies | | Descriptor: | Fab 1112-1 heavy chain variable domain, Fab 1112-1 light chain variable domain, Fab 17C7 heavy chain variable domain, ... | | Authors: | Ng, W.M, Fedosyuk, S, English, S, Augusto, G, Berg, A, Thorley, L, Haselon, A.S, Segireddy, R.R, Bowden, T.A, Douglas, A.D. | | Deposit date: | 2022-06-01 | | Release date: | 2022-08-17 | | Last modified: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structure of trimeric pre-fusion rabies virus glycoprotein in complex with two protective antibodies.
Cell Host Microbe, 30, 2022
|
|
5FPQ
 
 | | Structure of Homo sapiens acetylcholinesterase phosphonylated by sarin. | | Descriptor: | ACETYLCHOLINESTERASE, PENTAETHYLENE GLYCOL | | Authors: | Allgardsson, A, Berg, L, Akfur, C, Hornberg, A, Worek, F, Linusson, A, Ekstrom, F. | | Deposit date: | 2015-12-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Prereaction Complex between the Nerve Agent Sarin, its Biological Target Acetylcholinesterase, and the Antidote Hi-6.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5FPP
 
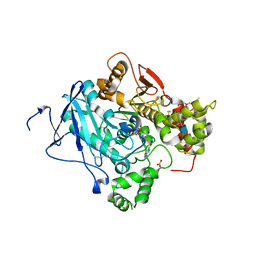 | | Structure of a pre-reaction ternary complex between sarin- acetylcholinesterase and HI-6 | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(AMINOCARBONYL)-1-[({2-[(E)-(HYDROXYIMINO)METHYL]PYRIDINIUM-1-YL}METHOXY)METHYL]PYRIDINIUM, ... | | Authors: | Allgardsson, A, Berg, L, Akfur, C, Hornberg, A, Worek, F, Linusson, A, Ekstrom, F. | | Deposit date: | 2015-12-02 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a Prereaction Complex between the Nerve Agent Sarin, its Biological Target Acetylcholinesterase, and the Antidote Hi-6.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4A23
 
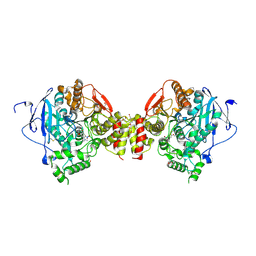 | | Mus musculus Acetylcholinesterase in complex with racemic C5685 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-(DIMETHYLAMINO)-N-{[(2R)-1-ETHYLPYRROLIDIN-2-YL]METHYL}-2-METHOXY-5-NITROBENZAMIDE, 4-(DIMETHYLAMINO)-N-{[(2S)-1-ETHYLPYRROLIDIN-2-YL]METHYL}-2-METHOXY-5-NITROBENZAMIDE, ... | | Authors: | Berg, L, Andersson, C.D, Artrursson, E, Hornberg, A, Tunemalm, A.K, Linusson, A, Ekstrom, F. | | Deposit date: | 2011-09-22 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting Acetylcholinesterase: Identification of Chemical Leads by High Throughput Screening, Structure Determination and Molecular Modeling.
Plos One, 6, 2011
|
|
4BBA
 
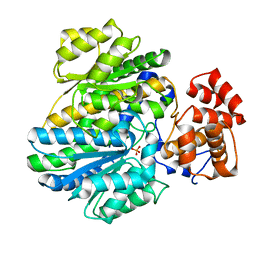 | | Crystal structure of glucokinase regulatory protein complexed to phosphate | | Descriptor: | GLUCOKINASE REGULATORY PROTEIN, PHOSPHATE ION | | Authors: | Pautsch, A, Stadler, N, Loehle, A, Lenter, M, Rist, W, Berg, A, Glocker, L, Nar, H, Reinhart, D, Heckel, A, Schnapp, G, Kauschke, S.G. | | Deposit date: | 2012-09-21 | | Release date: | 2013-05-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal Structure of Glucokinase Regulatory Protein.
Biochemistry, 52, 2013
|
|
4BB9
 
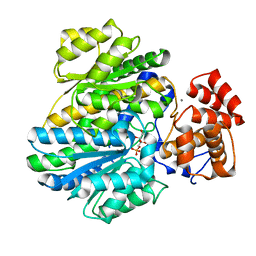 | | Crystal structure of glucokinase regulatory protein complexed to fructose-1-phosphate | | Descriptor: | 1-O-phosphono-beta-D-fructopyranose, CALCIUM ION, GLUCOKINASE REGULATORY PROTEIN | | Authors: | Pautsch, A, Stadler, N, Loehle, A, Lenter, M, Rist, W, Berg, A, Glocker, L, Nar, H, Reinert, D, Heckel, A, Schnapp, G, Kauschke, S.G. | | Deposit date: | 2012-09-21 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystal Structure of Glucokinase Regulatory Protein.
Biochemistry, 52, 2013
|
|
6GF1
 
 | | The structure of the ubiquitin-like modifier FAT10 reveals a novel targeting mechanism for degradation by the 26S proteasome | | Descriptor: | SULFATE ION, Ubiquitin D | | Authors: | Aichem, A, Anders, S, Catone, N, Roessler, P, Stotz, S, Berg, A, Schwab, R, Scheuermann, S, Bialas, J, Schmidtke, G, Peter, C, Groettrup, M, Wiesner, S. | | Deposit date: | 2018-04-28 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | The structure of the ubiquitin-like modifier FAT10 reveals an alternative targeting mechanism for proteasomal degradation.
Nat Commun, 9, 2018
|
|
