5M6Q
 
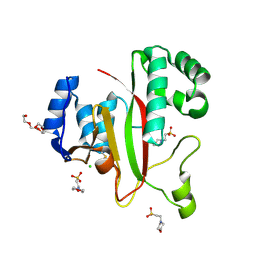 | | Crystal Structure of Kutzneria albida transglutaminase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Steffen, W, Benz, J, Rudolph, M.G. | | Deposit date: | 2016-10-25 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of a microbial transglutaminase enabling highly site-specific labeling of proteins.
J. Biol. Chem., 292, 2017
|
|
5MQY
 
 | | CATHEPSIN L IN COMPLEX WITH 4-[1,3-benzodioxol-5-ylmethyl(2-phenoxyethyl)amino]-5-fluoropyrimidine-2-carbonitrile | | Descriptor: | 1,2-ETHANEDIOL, 4-[1,3-benzodioxol-5-ylmethyl(2-phenoxyethyl)amino]-5-fluoropyrimidine-2-carbonitrile, Cathepsin L1 | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-12-21 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Prospective Evaluation of Free Energy Calculations for the Prioritization of Cathepsin L Inhibitors.
J. Med. Chem., 60, 2017
|
|
5N89
 
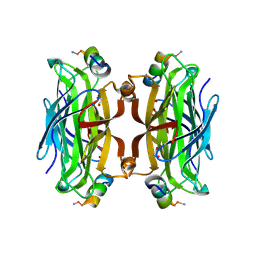 | | CRYSTAL STRUCTURE OF STREPTAVIDIN WITH PEPTIDE GNSFDDWLASKG | | Descriptor: | GLY-ASN-SER-PHE-ASP-ASP-TRP-LEU-ALA-SER-LYS-GLY-NH2, GLYCEROL, Streptavidin | | Authors: | Lyamichev, V, Goodrich, L, Sullivan, E, Bannen, R, Benz, J, Albert, T, Patel, J. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Stepwise Evolution Improves Identification of Diverse Peptides Binding to a Protein Target.
Sci Rep, 7, 2017
|
|
5N8W
 
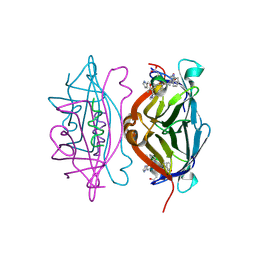 | | CRYSTAL STRUCTURE OF STREPTAVIDIN with D-amino acid containing peptide GGwhdeatwkpG | | Descriptor: | GLY-GLY-DTR-DHI-DAS-DGL-DAL-DTH-DTR-DLY, Streptavidin | | Authors: | Lyamichev, V, Goodrich, L, Sullivan, E, Bannen, R, Benz, J, Albert, T, Patel, J. | | Deposit date: | 2017-02-24 | | Release date: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Stepwise Evolution Improves Identification of Diverse Peptides Binding to a Protein Target.
Sci Rep, 7, 2017
|
|
5N8T
 
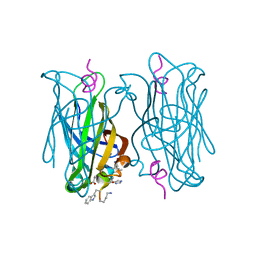 | | CRYSTAL STRUCTURE OF STREPTAVIDIN D-amino acid containing peptide Gdlwqheatwkkq | | Descriptor: | DLE-DTR-DGN-DHI-DGL-DAL-DTH-DTR-DLY, Streptavidin | | Authors: | Lyamichev, V, Goodrich, L, Sullivan, E, Bannen, R, Benz, J, Albert, T, Patel, J. | | Deposit date: | 2017-02-24 | | Release date: | 2017-10-04 | | Last modified: | 2022-12-07 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Stepwise Evolution Improves Identification of Diverse Peptides Binding to a Protein Target.
Sci Rep, 7, 2017
|
|
5N8E
 
 | | CRYSTAL STRUCTURE OF STREPTAVIDIN WITH PEPTIDE RDPAPAWAHGGG | | Descriptor: | ARG-ASP-PRO-ALA-PRO-ALA-TRP-ALA-HIS-GLY-GLY-GLY-NH2, SODIUM ION, Streptavidin | | Authors: | Lyamichev, V, Goodrich, L, Sullivan, E, Bannen, R, Benz, J, Albert, T, Patel, J. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Stepwise Evolution Improves Identification of Diverse Peptides Binding to a Protein Target.
Sci Rep, 7, 2017
|
|
5N99
 
 | | CRYSTAL STRUCTURE OF STREPTAVIDIN with cyclic peptide NQpWQ | | Descriptor: | ASN-GLN-DPR-TRP-GLN, Streptavidin | | Authors: | Lyamichev, V, Goodrich, L, Sullivan, E, Bannen, R, Benz, J, Albert, T, Patel, J. | | Deposit date: | 2017-02-24 | | Release date: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Stepwise Evolution Improves Identification of Diverse Peptides Binding to a Protein Target.
Sci Rep, 7, 2017
|
|
5N8J
 
 | | CRYSTAL STRUCTURE OF STREPTAVIDIN WITH PEPTIDE D-amino acid containing peptide GyGlanvdessG | | Descriptor: | GLY-DTY-GLY-DLE-DAL-DSG-DVA-DAS-DGL-DSN-DSN-GLY, ISOPROPYL ALCOHOL, Streptavidin | | Authors: | Lyamichev, V, Goodrich, L, Sullivan, E, Bannen, R, Benz, J, Albert, T, Patel, J. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Stepwise Evolution Improves Identification of Diverse Peptides Binding to a Protein Target.
Sci Rep, 7, 2017
|
|
5N7X
 
 | | CRYSTAL STRUCTURE OF STREPTAVIDIN WITH PEPTIDE EWVHPQFEQKAK | | Descriptor: | GLU-TRP-VAL-HIS-PRO-GLN-PHE-GLU-GLN-LYS-ALA-LYS Peptide, Streptavidin | | Authors: | Lyamichev, V, Goodrich, L, Sullivan, E, Bannen, R, Benz, J, Albert, T, Patel, J. | | Deposit date: | 2017-02-21 | | Release date: | 2017-10-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Stepwise Evolution Improves Identification of Diverse Peptides Binding to a Protein Target.
Sci Rep, 7, 2017
|
|
5N8B
 
 | | CRYSTAL STRUCTURE OF STREPTAVIDIN WITH PEPTIDE AFPDYLAEYHGG | | Descriptor: | ALA-PHE-PRO-ASP-TYR-LEU-ALA-GLU-TYR-HIS-GLY-GLY-NH2, Streptavidin | | Authors: | Lyamichev, V, Goodrich, L, Sullivan, E, Bannen, R, Benz, J, Albert, T, Patel, J. | | Deposit date: | 2017-02-23 | | Release date: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Stepwise Evolution Improves Identification of Diverse Peptides Binding to a Protein Target.
Sci Rep, 7, 2017
|
|
5M5E
 
 | | Crystal structure of a interleukin-2 variant in complex with interleukin-2 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Klein, C, Freimoser-Grundschober, A, Waldhauer, I, Stihle, M, Birk, M, Benz, J. | | Deposit date: | 2016-10-21 | | Release date: | 2017-05-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cergutuzumab amunaleukin (CEA-IL2v), a CEA-targeted IL-2 variant-based immunocytokine for combination cancer immunotherapy: Overcoming limitations of aldesleukin and conventional IL-2-based immunocytokines.
Oncoimmunology, 6, 2017
|
|
5MCQ
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH ACTIVE SITE AND EXOSITE BINDING PEPTIDE INHIBITOR | | Descriptor: | 1,2-ETHANEDIOL, BACE-1 ACTIVE AND EXOSITE BINDING INHIBITOR, Beta-secretase 1 | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
5MBW
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH Pep#3 | | Descriptor: | BACE1 INHIBITOR PEPTIDE Pep#3, Beta-secretase 1, CHLORIDE ION | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-09 | | Release date: | 2017-09-27 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
5MCO
 
 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH ACTIVE SITE INHIBITOR GRL-8234 AND EXOSITE PEPTIDE | | Descriptor: | BACE-1 EXOSITE PEPTIDE, Beta-secretase 1, N-{(1S,2R)-1-benzyl-2-hydroxy-3-[(3-methoxybenzyl)amino]propyl}-5-[methyl(methylsulfonyl)amino]-N'-[(1R)-1-phenylethyl]benzene-1,3-dicarboxamide | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-09-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Potent and Selective BACE-1 Peptide Inhibitors Lower Brain A beta Levels Mediated by Brain Shuttle Transport.
EBioMedicine, 24, 2017
|
|
5MAJ
 
 | | CATHEPSIN L IN COMPLEX WITH 4-[cyclopentyl(imidazo[1,2-a]pyridin-2-ylmethyl)amino]-6-morpholino-1,3,5-triazine-2-carbonitrile | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L1, ~{N}-cyclopentyl-~{N}-(imidazo[1,2-a]pyridin-2-ylmethyl)-4-(iminomethyl)-6-morpholin-4-yl-1,3,5-triazin-2-amine | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Inhibition of the Cysteine Protease Human Cathepsin L by Triazine Nitriles: AmideHeteroarene pi-Stacking Interactions and Chalcogen Bonding in the S3 Pocket.
ChemMedChem, 12, 2017
|
|
5MAE
 
 | | CATHEPSIN L IN COMPLEX WITH (2S,4R)-4-(2-Chloro-4-methoxy-benzenesulfonyl)-1-[3-(5-chloro-pyridin-2-yl)-azetidine-3-carbonyl]-pyrrolidine-2-car boxylic acid (1-cyano-cyclopropyl)-amide | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin L1, [4-[cyclopentyl(pyrazin-2-ylmethyl)amino]-6-morpholin-4-yl-1,3,5-triazin-2-yl]methylideneazanide | | Authors: | Kuglstatter, A, Stihle, M, Benz, J. | | Deposit date: | 2016-11-03 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Inhibition of the Cysteine Protease Human Cathepsin L by Triazine Nitriles: AmideHeteroarene pi-Stacking Interactions and Chalcogen Bonding in the S3 Pocket.
ChemMedChem, 12, 2017
|
|
3ODI
 
 | | Crystal structure of cyclophilin A in complex with Voclosporin E-ISA247 | | Descriptor: | Cyclophilin A, Voclosporin | | Authors: | Kuglstatter, A, Stihle, M, Benz, J, Hennig, M. | | Deposit date: | 2010-08-11 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for the cyclophilin A binding affinity and immunosuppressive potency of E-ISA247 (voclosporin).
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3S3X
 
 | | Structure of chicken acid-sensing ion channel 1 AT 3.0 A resolution in complex with psalmotoxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Amiloride-sensitive cation channel 2, neuronal, ... | | Authors: | Dawson, R.J.P, Benz, J, Stohler, P, Tetaz, T, Joseph, C, Huber, S, Schmid, G, Huegin, D, Pflimlin, P, Trube, G, Rudolph, M.G, Hennig, M, Ruf, A. | | Deposit date: | 2011-05-18 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structure of the Acid-sensing ion channel 1 in complex with the gating modifier Psalmotoxin 1.
Nat Commun, 3, 2012
|
|
3S3W
 
 | | Structure of chicken acid-sensing ion channel 1 at 2.6 a resolution and ph 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Amiloride-sensitive cation channel 2, ... | | Authors: | Dawson, R.J.P, Benz, J, Stohler, P, Tetaz, T, Joseph, C, Huber, S, Schmid, G, Huegin, D, Pflimlin, P, Trube, G, Rudolph, M.G, Hennig, M, Ruf, A. | | Deposit date: | 2011-05-18 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Acid-sensing ion channel 1 in complex with the gating modifier Psalmotoxin 1.
Nat Commun, 3, 2012
|
|
3ODL
 
 | | Crystal structure of cyclophilin A in complex with Voclosporin Z-ISA247 | | Descriptor: | Cyclophilin A, Voclosporin | | Authors: | Kuglstatter, A, Stihle, M, Benz, J, Hennig, M. | | Deposit date: | 2010-08-11 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis for the cyclophilin A binding affinity and immunosuppressive potency of E-ISA247 (voclosporin).
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3SGJ
 
 | | Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgIII and antibodies lacking core fucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MALONATE ION, ... | | Authors: | Ferrara, C, Grau, S, Jaeger, C, Sondermann, P, Bruenker, P, Waldhauer, I, Hennig, M, Ruf, A, Rufer, A.C, Stihle, M, Umana, P, Benz, J. | | Deposit date: | 2011-06-15 | | Release date: | 2011-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4P7F
 
 | | Mouse apo-COMT | | Descriptor: | Catechol O-methyltransferase, HYDROGENPHOSPHATE ION, PHOSPHATE ION, ... | | Authors: | Ehler, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Mapping the conformational space accessible to catechol-O-methyltransferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4P7K
 
 | | Rat COMT in complex with sinefungin | | Descriptor: | ACETATE ION, Catechol O-methyltransferase, L(+)-TARTARIC ACID, ... | | Authors: | Ehler, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Mapping the conformational space accessible to catechol-O-methyltransferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4P7G
 
 | | Rat apo-COMT, phosphate bound | | Descriptor: | Catechol O-methyltransferase, PHOSPHATE ION, POTASSIUM ION | | Authors: | Ehler, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Mapping the conformational space accessible to catechol-O-methyltransferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4PYJ
 
 | | human apo-COMT, single domain swap | | Descriptor: | CHLORIDE ION, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Ehler, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mapping the conformational space accessible to catechol-O-methyltransferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
