1QPL
 
 | | FK506 BINDING PROTEIN (12 KDA, HUMAN) COMPLEX WITH L-707,587 | | Descriptor: | C32-O-(1-METHYL-INDOL-5-YL) 18-HYDROXY-ASCOMYCIN, PROTEIN (FK506-BINDING PROTEIN) | | Authors: | Becker, J.W, Rotonda, J. | | Deposit date: | 1999-05-25 | | Release date: | 1999-08-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | 32-Indolyl ether derivatives of ascomycin: three-dimensional structures of complexes with FK506-binding protein.
J.Med.Chem., 42, 1999
|
|
1RE1
 
 | |
1BMG
 
 | |
2FKE
 
 | | FK-506-BINDING PROTEIN: THREE-DIMENSIONAL STRUCTURE OF THE COMPLEX WITH THE ANTAGONIST L-685,818 | | Descriptor: | 8-DEETHYL-8-[BUT-3-ENYL]-ASCOMYCIN, FK506 BINDING PROTEIN | | Authors: | Becker, J.W, Mckeever, B.M, Rotonda, J. | | Deposit date: | 1993-01-27 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | FK-506-binding protein: three-dimensional structure of the complex with the antagonist L-685,818.
J.Biol.Chem., 268, 1993
|
|
1HFS
 
 | |
1IBC
 
 | |
1FKD
 
 | | FK-506 BINDING PROTEIN: THREE-DIMENSIONAL STRUCTURE OF THE COMPLEX WITH THE ANTAGONIST L-685,818 | | Descriptor: | 18-HYDROXYASCOMYCIN, FK506 BINDING PROTEIN | | Authors: | Becker, J.W, Rotonda, J, Mckeever, B.M. | | Deposit date: | 1992-12-02 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | FK-506-binding protein: three-dimensional structure of the complex with the antagonist L-685,818.
J.Biol.Chem., 268, 1993
|
|
1SLM
 
 | |
1SLN
 
 | |
1RHJ
 
 | |
1RHU
 
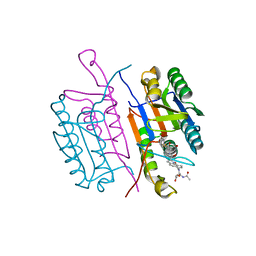 | | CRYSTAL STRUCTURE OF THE COMPLEX OF CASPASE-3 WITH A 5,6,7 TRICYCLIC PEPTIDOMIMETIC INHIBITOR | | Descriptor: | (3S)-3-[({(2S)-5-[(N-ACETYL-L-ALPHA-ASPARTYL)AMINO]-4-OXO-1,2,4,5,6,7-HEXAHYDROAZEPINO[3,2,1-HI]INDOL-2-YL}CARBONYL)AMINO]-5-(BENZYLSULFANYL)-4-OXOPENTANOIC ACID, Caspase-3 | | Authors: | Becker, J.W, Rotonda, J, Soisson, S.M. | | Deposit date: | 2003-11-14 | | Release date: | 2004-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Reducing the Peptidyl Features of Caspase-3 Inhibitors: A Structural Analysis.
J.Med.Chem., 47, 2004
|
|
1RHQ
 
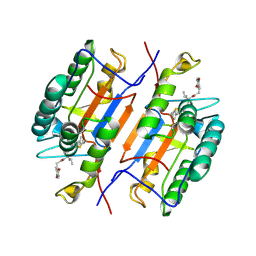 | |
1RHM
 
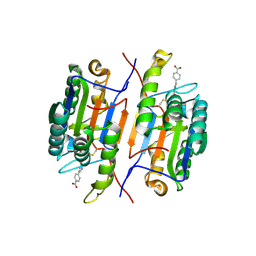 | |
1RHK
 
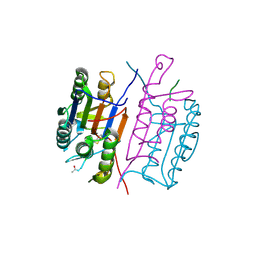 | |
1RHR
 
 | |
1QPF
 
 | | FK506 BINDING PROTEIN (12 KDA, HUMAN) COMPLEX WITH L-709,858 | | Descriptor: | C32-O-(1-ETHYL-INDOL-5-YL)ASCOMYCIN, PROTEIN (FK506-BINDING PROTEIN), heptyl beta-D-glucopyranoside | | Authors: | Becker, J.W, Rotonda, J. | | Deposit date: | 1999-05-24 | | Release date: | 1999-08-16 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 32-Indolyl ether derivatives of ascomycin: three-dimensional structures of complexes with FK506-binding protein.
J.Med.Chem., 42, 1999
|
|
1Q6T
 
 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 11 | | Descriptor: | 6-[4-((2S)-2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-3-{4-[DIFLUORO(PHOSPHONO)METHYL]PHENYL}-2-PHENYLPROPYL)PHENYL]-2-[(1S)-1-METHOXY-3-METHYLBUTYL]QUINOLIN-8-YLPHOSPHONIC ACID, MAGNESIUM ION, Protein-tyrosine phosphatase, ... | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
1Q6S
 
 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 9 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-[4-((2R)-2-(1H-1,2,3-BENZOTRIAZOL-1-YL)-3-{4-[DIFLUORO(PHOSPHONO)METHYL]PHENYL}-2-PHENYLPROPYL)PHENYL]-2-METHYLQUINOLIN-8-YLPHOSPHONIC ACID, CHLORIDE ION, ... | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
1Q6N
 
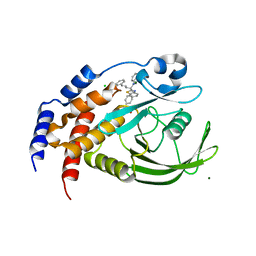 | | THE STRUCTURE OF PHOSPHOTYROSINE PHOSPHATASE 1B IN COMPLEX WITH COMPOUND 4 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Protein-tyrosine phosphatase, ... | | Authors: | Scapin, G, Patel, S.B, Becker, J.W, Wang, Q, Desponts, C, Waddleton, D, Skorey, K, Cromlish, W, Bayly, C, Therien, M, Gauthier, J.Y, Li, C.S, Lau, C.K, Ramachandran, C, Kennedy, B.P, Asante-Appiah, E. | | Deposit date: | 2003-08-13 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structural Basis for the Selectivity of Benzotriazole Inhibitors of Ptp1B
Biochemistry, 42, 2003
|
|
4Z9L
 
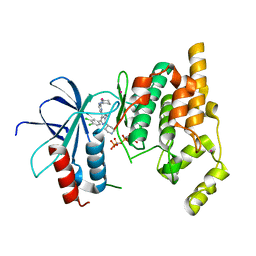 | | THE STRUCTURE OF JNK3 IN COMPLEX WITH AN IMIDAZOLE-PYRIMIDINE INHIBITOR | | Descriptor: | CYCLOHEXYL-{4-[5-(3,4-DICHLOROPHENYL)-2-PIPERIDIN-4-YL-3-PROPYL-3H-IMIDAZOL-4-YL]-PYRIMIDIN-2-YL}AMINE, Mitogen-activated protein kinase 10, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Scapin, G, Patel, S.B, Lisnock, J, Becker, J.W, Lograsso, P.V, Smart, O.S, Bricogne, G. | | Deposit date: | 2015-04-10 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of JNK3 in complex with small molecule inhibitors: structural basis for potency and selectivity.
Chem.Biol., 10, 2003
|
|
2B7Y
 
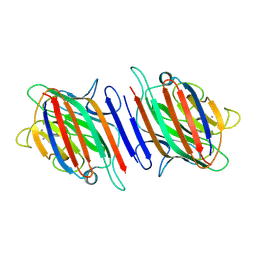 | | Fava Bean Lectin-Glucose Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Favin alpha chain, ... | | Authors: | Reeke Jr, G.N, Becker, J.W. | | Deposit date: | 2005-10-05 | | Release date: | 2005-10-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structure of favin: saccharide binding-cyclic permutation in leguminous lectins
Science, 234, 1986
|
|
2CNA
 
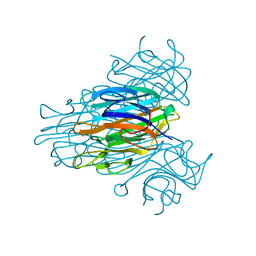 | | THE COVALENT AND THREE-DIMENSIONAL STRUCTURE OF CONCANAVALIN A, IV.ATOMIC COORDINATES,HYDROGEN BONDING,AND QUATERNARY STRUCTURE | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION | | Authors: | Reekejunior, G.N, Becker, J.W, Edelman, G.M. | | Deposit date: | 1975-04-01 | | Release date: | 1977-03-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The covalent and three-dimensional structure of concanavalin A. IV. Atomic coordinates, hydrogen bonding, and quaternary structure.
J.Biol.Chem., 250, 1975
|
|
3BEJ
 
 | | Structure of human FXR in complex with MFA-1 and co-activator peptide | | Descriptor: | (8alpha,10alpha,13alpha,17beta)-17-[(4-hydroxyphenyl)carbonyl]androsta-3,5-diene-3-carboxylic acid, Bile acid receptor, Nuclear receptor coactivator 1, ... | | Authors: | Soisson, S.M, Parthasarathy, G, Becker, J.W. | | Deposit date: | 2007-11-19 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Identification of a potent synthetic FXR agonist with an unexpected mode of binding and activation.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1YAT
 
 | |
1IAU
 
 | | HUMAN GRANZYME B IN COMPLEX WITH AC-IEPD-CHO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, GRANZYME B, ... | | Authors: | Rotonda, J, Garcia-Calvo, M, Bull, H.G, Geissler, W.M, McKeever, B.M, Willoughby, C.A, Thornberry, N.A, Becker, J.W. | | Deposit date: | 2001-03-23 | | Release date: | 2001-05-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The three-dimensional structure of human granzyme B compared to caspase-3, key mediators of cell death with cleavage specificity for aspartic acid in P1.
Chem.Biol., 8, 2001
|
|
