1A5Y
 
 | |
1FQ1
 
 | | CRYSTAL STRUCTURE OF KINASE ASSOCIATED PHOSPHATASE (KAP) IN COMPLEX WITH PHOSPHO-CDK2 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN KINASE 2, CYCLIN-DEPENDENT KINASE INHIBITOR 3, ... | | Authors: | Song, H, Hanlon, N, Brown, N.R, Noble, M.E.M, Johnson, L.N, Barford, D. | | Deposit date: | 2000-09-01 | | Release date: | 2001-05-09 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Phosphoprotein-protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2.
Mol.Cell, 7, 2001
|
|
1ABB
 
 | | CONTROL OF PHOSPHORYLASE B CONFORMATION BY A MODIFIED COFACTOR: CRYSTALLOGRAPHIC STUDIES ON R-STATE GLYCOGEN PHOSPHORYLASE RECONSTITUTED WITH PYRIDOXAL 5'-DIPHOSPHATE | | Descriptor: | GLYCOGEN PHOSPHORYLASE B, INOSINIC ACID, PYRIDOXAL-5'-DIPHOSPHATE, ... | | Authors: | Leonidas, D.D, Oikonomakos, N.G, Papageorgiou, A.C, Acharya, K.R, Barford, D, Johnson, L.N. | | Deposit date: | 1992-04-09 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Control of phosphorylase b conformation by a modified cofactor: crystallographic studies on R-state glycogen phosphorylase reconstituted with pyridoxal 5'-diphosphate.
Protein Sci., 1, 1992
|
|
1G1G
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH A MONO-PHOSPHORYLATED PEPTIDE (ETDY(PTR)RKGGKGLL) FROM THE INSULIN RECEPTOR KINASE | | Descriptor: | MONO-PHOSPHORYLATED PEPTIDE FROM THE INSULIN RECEPTOR KINASE, PROTEIN TYROSINE PHOSPHATASE 1B | | Authors: | Salmeen, A, Andersen, J.N, Myers, M.P, Tonks, N.K, Barford, D. | | Deposit date: | 2000-10-11 | | Release date: | 2001-01-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B.
Mol.Cell, 6, 2000
|
|
1G1F
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH A TRI-PHOSPHORYLATED PEPTIDE (RDI(PTR)ETD(PTR)(PTR)RK) FROM THE INSULIN RECEPTOR KINASE | | Descriptor: | PROTEIN TYROSINE PHOSPHATASE 1B, TRI-PHOSPHORYLATED PEPTIDE FROM THE INSULIN RECEPTOR KINASE | | Authors: | Salmeen, A, Andersen, J.N, Myers, M.P, Tonks, N.K, Barford, D. | | Deposit date: | 2000-10-11 | | Release date: | 2001-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B.
Mol.Cell, 6, 2000
|
|
1G1H
 
 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH A BIS-PHOSPHORYLATED PEPTIDE (ETD(PTR)(PTR)RKGGKGLL) FROM THE INSULIN RECEPTOR KINASE | | Descriptor: | BI-PHOSPHORYLATED PEPTIDE FROM THE INSULIN RECEPTOR KINASE, PROTEIN TYROSINE PHOSPHATASE 1B | | Authors: | Salmeen, A, Andersen, J.N, Myers, M.P, Tonks, N.K, Barford, D. | | Deposit date: | 2000-10-11 | | Release date: | 2001-01-17 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B.
Mol.Cell, 6, 2000
|
|
1OEO
 
 | | PTP1B with the catalytic cysteine oxidized to sulfonic acid | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | Salmeen, A, Andersen, J.N, Myers, M.P, Meng, T.C, Hinks, J.A, Tonks, N.K, Barford, D. | | Deposit date: | 2003-03-28 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Redox Regulation of Protein Tyrosine Phosphatase Involves a Sulfenyl-Amide Intermediate
Nature, 423, 2003
|
|
1FPZ
 
 | | CRYSTAL STRUCTURE ANALYSIS OF KINASE ASSOCIATED PHOSPHATASE (KAP) WITH A SUBSTITUTION OF THE CATALYTIC SITE CYSTEINE (CYS140) TO A SERINE | | Descriptor: | CYCLIN-DEPENDENT KINASE INHIBITOR 3, SULFATE ION | | Authors: | Song, H, Hanlon, N, Brown, N.R, Noble, M.E.M, Johnson, L.N, Barford, D. | | Deposit date: | 2000-09-01 | | Release date: | 2001-05-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphoprotein-protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2.
Mol.Cell, 7, 2001
|
|
1OEM
 
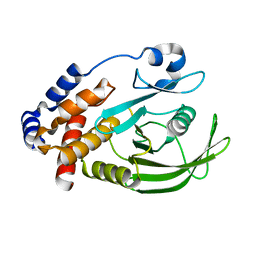 | | PTP1B with the catalytic cysteine oxidized to a sulfenyl-amide bond | | Descriptor: | PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | Salmeen, A, Andersen, J.N, Myers, M.P, Meng, T.C, Hinks, J.A, Tonks, N.K, Barford, D. | | Deposit date: | 2003-03-28 | | Release date: | 2003-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Redox Regulation of Protein Tyrosine Phosphatase Involves a Sulfenyl-Amide Intermediate
Nature, 423, 2003
|
|
2IXM
 
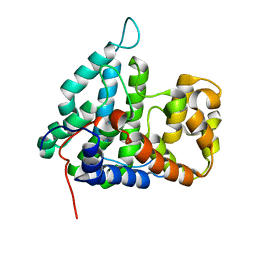 | | Structure of human PTPA | | Descriptor: | SERINE/THREONINE-PROTEIN PHOSPHATASE 2A REGULATORY SUBUNIT B' | | Authors: | Leulliot, N, Vicentini, G, Jordens, J, Quevillon-Cheruel, S, Schiltz, M, Barford, D, Van Tilbeurgh, H, Goris, J. | | Deposit date: | 2006-07-09 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the Pp2A Phosphatase Activator: Implications for its Pp2A-Specific Ppiase Activity
Mol.Cell, 23, 2006
|
|
2IXN
 
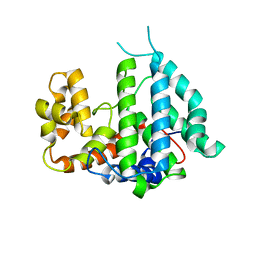 | | CRYSTAL STRUCTURE OF THE PP2A PHOSPHATASE ACTIVATOR Ypa2 PTPA2 | | Descriptor: | SERINE/THREONINE-PROTEIN PHOSPHATASE 2A ACTIVATOR 2 | | Authors: | Leulliot, N, Vicentini, G, Jordens, J, Quevillon-Cheruel, S, Schiltz, M, Barford, D, Van Tilbeurgh, H, Goris, J. | | Deposit date: | 2006-07-09 | | Release date: | 2006-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Pp2A Phosphatase Activator: Implications for its Pp2A-Specific Ppiase Activity
Mol.Cell, 23, 2006
|
|
1A6Q
 
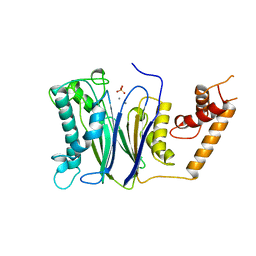 | | CRYSTAL STRUCTURE OF THE PROTEIN SERINE/THREONINE PHOSPHATASE 2C AT 2 A RESOLUTION | | Descriptor: | MANGANESE (II) ION, PHOSPHATASE 2C, PHOSPHATE ION | | Authors: | Das, A.K, Helps, N.R, Cohen, P.T.W, Barford, D. | | Deposit date: | 1998-02-27 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the protein serine/threonine phosphatase 2C at 2.0 A resolution.
EMBO J., 15, 1996
|
|
2IXO
 
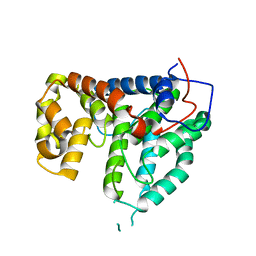 | | CRYSTAL STRUCTURE OF THE PP2A PHOSPHATASE ACTIVATOR Ypa1 PTPA1 | | Descriptor: | SERINE/THREONINE-PROTEIN PHOSPHATASE 2A ACTIVATOR 1 | | Authors: | Leulliot, N, Vicentini, G, Jordens, J, Quevillon-Cheruel, S, Schiltz, M, Barford, D, Van Tilbeurgh, H, Goris, J. | | Deposit date: | 2006-07-09 | | Release date: | 2006-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Pp2A Phosphatase Activator: Implications for its Pp2A-Specific Ppiase Activity.
Mol.Cell, 23, 2006
|
|
1B3U
 
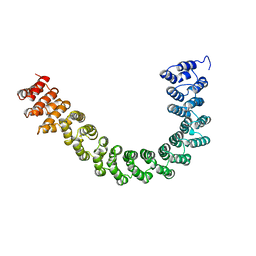 | | CRYSTAL STRUCTURE OF CONSTANT REGULATORY DOMAIN OF HUMAN PP2A, PR65ALPHA | | Descriptor: | PROTEIN (PROTEIN PHOSPHATASE PP2A) | | Authors: | Groves, M.R, Hanlon, N, Turowski, P, Hemmings, B, Barford, D. | | Deposit date: | 1998-12-14 | | Release date: | 1999-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs.
Cell(Cambridge,Mass.), 96, 1999
|
|
1H6G
 
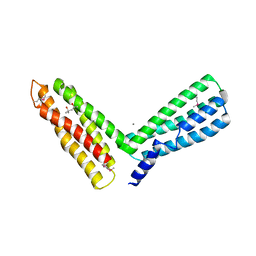 | | alpha-catenin M-domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ALPHA-1 CATENIN, CALCIUM ION, ... | | Authors: | Yang, J, Dokurno, P, Tonks, N.K, Barford, D. | | Deposit date: | 2001-06-14 | | Release date: | 2001-08-07 | | Last modified: | 2016-02-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the M-Fragment of Alpha-Catenin: Implications for Modulation of Cell Adhesion.
Embo J., 20, 2001
|
|
2IXP
 
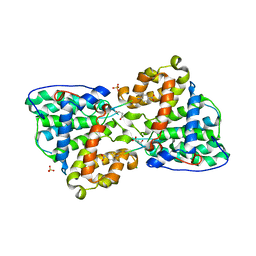 | | Crystal structure of the Pp2A phosphatase activator Ypa1 PTPA1 in complex with model substrate | | Descriptor: | CHLORIDE ION, SERINE/THREONINE-PROTEIN PHOSPHATASE 2A ACTIVATOR 1, SIN-ALA-ALA-PRO-LYS-NIT, ... | | Authors: | Leulliot, N, Vicentini, G, Jordens, J, Quevillon-Cheruel, S, Schiltz, M, Barford, D, Van Tilbeurgh, H, Goris, J. | | Deposit date: | 2006-07-09 | | Release date: | 2006-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the PP2A phosphatase activator: implications for its PP2A-specific PPIase activity.
Mol. Cell, 23, 2006
|
|
2JDO
 
 | | STRUCTURE OF PKB-BETA (AKT2) COMPLEXED WITH ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY) ETHYLAMINO)ETHYL)AMIDE | | Descriptor: | 1,2-ETHANEDIOL, GLYCOGEN SYNTHASE KINASE-3 BETA, ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY)ETHYLAMINO)ETHYL)AMIDE, ... | | Authors: | Davies, T.G, Verdonk, M.L, Graham, B, Saalau-Bethell, S, Hamlett, C.C.F, Mchardy, T, Collins, I, Garrett, M.D, Workman, P, Woodhead, S.J, Jhoti, H, Barford, D. | | Deposit date: | 2007-01-11 | | Release date: | 2007-02-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Structural Comparison of Inhibitor Binding to Pkb, Pka and Pka-Pkb Chimera
J.Mol.Biol., 367, 2007
|
|
2JDR
 
 | | STRUCTURE OF PKB-BETA (AKT2) COMPLEXED WITH THE INHIBITOR A-443654 | | Descriptor: | (2S)-1-(1H-INDOL-3-YL)-3-{[5-(3-METHYL-1H-INDAZOL-5-YL)PYRIDIN-3-YL]OXY}PROPAN-2-AMINE, GLYCOGEN SYNTHASE KINASE-3 BETA, RAC-BETA SERINE/THREONINE-PROTEIN KINASE | | Authors: | Davies, T.G, Verdonk, M.L, Graham, B, Saalau-Bethell, S, Hamlett, C.C.F, McHardy, T, Collins, I, Garrett, M.D, Workman, P, Woodhead, S.J, Jhoti, H, Barford, D. | | Deposit date: | 2007-01-12 | | Release date: | 2007-02-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Structural Comparison of Inhibitor Binding to Pkb, Pka and Pka-Pkb Chimera
J.Mol.Biol., 367, 2007
|
|
2JDV
 
 | | Structure of PKA-PKB chimera complexed with A-443654 | | Descriptor: | (2S)-1-(1H-INDOL-3-YL)-3-{[5-(3-METHYL-1H-INDAZOL-5-YL)PYRIDIN-3-YL]OXY}PROPAN-2-AMINE, CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA | | Authors: | Davies, T.G, Verdonk, M.L, Graham, B, Saalau-Bethell, S, Hamlett, C.C.F, Mchardy, T, Collins, I, Garrett, M.D, Workman, P, Woodhead, S.J, Jhoti, H, Barford, D. | | Deposit date: | 2007-01-12 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A Structural Comparison of Inhibitor Binding to Pkb, Pka and Pka-Pkb Chimera
J.Mol.Biol., 367, 2007
|
|
2WMN
 
 | | Structure of the complex between DOCK9 and Cdc42-GDP. | | Descriptor: | CELL DIVISION CONTROL PROTEIN 42 HOMOLOG, DEDICATOR OF CYTOKINESIS PROTEIN 9, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Yang, J, Roe, S.M, Barford, D. | | Deposit date: | 2009-07-02 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Activation of Rho Gtpases by Dock Exchange Factors is Mediated by a Nucleotide Sensor.
Science, 325, 2009
|
|
2WM9
 
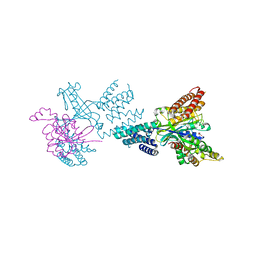 | | Structure of the complex between DOCK9 and Cdc42. | | Descriptor: | CELL DIVISION CONTROL PROTEIN 42 HOMOLOG, DEDICATOR OF CYTOKINESIS PROTEIN 9, GLYCEROL | | Authors: | Yang, J, Roe, S.M, Barford, D. | | Deposit date: | 2009-06-30 | | Release date: | 2009-09-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activation of Rho Gtpases by Dock Exchange Factors is Mediated by a Nucleotide Sensor.
Science, 325, 2009
|
|
2WMO
 
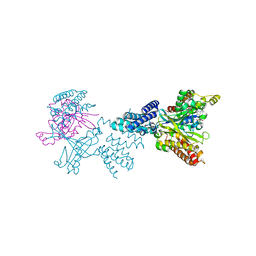 | | Structure of the complex between DOCK9 and Cdc42. | | Descriptor: | CELL DIVISION CONTROL PROTEIN 42 HOMOLOG, DEDICATOR OF CYTOKINESIS PROTEIN 9, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yang, J, Roe, S.M, Barford, D. | | Deposit date: | 2009-07-02 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Activation of Rho Gtpases by Dock Exchange Factors is Mediated by a Nucleotide Sensor.
Science, 325, 2009
|
|
1GQP
 
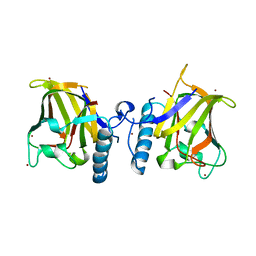 | | APC10/DOC1 SUBUNIT OF S. cerevisiae | | Descriptor: | BROMIDE ION, DOC1/APC10 | | Authors: | Au, S.W.N, Leng, X, Harper, J.W.A.D.E, Barford, D. | | Deposit date: | 2001-11-28 | | Release date: | 2002-03-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Implications for the Ubiquitination Reaction of the Anaphase-Promoting Complex from the Crystal Structure of the Doc1/Apc10 Subunit.
J.Mol.Biol., 316, 2002
|
|
2JDS
 
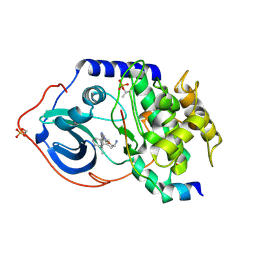 | | Structure of cAMP-dependent protein kinase complexed with A-443654 | | Descriptor: | (2S)-1-(1H-INDOL-3-YL)-3-{[5-(3-METHYL-1H-INDAZOL-5-YL)PYRIDIN-3-YL]OXY}PROPAN-2-AMINE, CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA | | Authors: | Davies, T.G, Verdonk, M.L, Graham, B, Saalau-Bethell, S, Hamlett, C.C.F, McHardy, T, Collins, I, Garrett, M.D, Workman, P, Woodhead, S.J, Jhoti, H, Barford, D. | | Deposit date: | 2007-01-12 | | Release date: | 2007-02-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural Comparison of Inhibitor Binding to Pkb, Pka and Pka-Pkb Chimera
J.Mol.Biol., 367, 2007
|
|
2JDT
 
 | | Structure of PKA-PKB chimera complexed with ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY) ETHYLAMINO)ETHYL)AMIDE | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY)ETHYLAMINO)ETHYL)AMIDE | | Authors: | Davies, T.G, Verdonk, M.L, Graham, B, Saalau-Bethell, S, Hamlett, C.C.F, McHardy, T, Collins, I, Garrett, M.D, Workman, P, Woodhead, S.J, Jhoti, H, Barford, D. | | Deposit date: | 2007-01-12 | | Release date: | 2007-02-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Structural Comparison of Inhibitor Binding to Pkb, Pka and Pka-Pkb Chimera
J.Mol.Biol., 367, 2007
|
|
