3UD6
 
 | | Structural analyses of covalent enzyme-substrate analogue complexes reveal strengths and limitations of de novo enzyme design | | Descriptor: | 1-(6-METHOXYNAPHTHALEN-2-YL)BUTANE-1,3-DIONE, RETRO-ALDOLASE, SULFATE ION | | Authors: | Baker, D, Stoddard, B.L, Althoff, E.A, Wang, L, Jiang, L, Moody, J, Bolduc, J, Lassila, J, Hilvert, D. | | Deposit date: | 2011-10-27 | | Release date: | 2011-11-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | Structural analyses of covalent enzyme-substrate analog complexes reveal strengths and limitations of de novo enzyme design.
J.Mol.Biol., 415, 2012
|
|
7M0Q
 
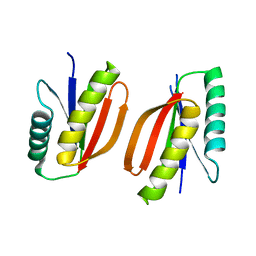 | |
2KL8
 
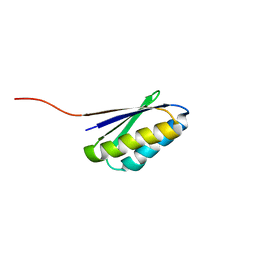 | | Solution NMR Structure of de novo designed ferredoxin-like fold protein, Northeast Structural Genomics Consortium Target OR15 | | Descriptor: | OR15 | | Authors: | Liu, G, Koga, N, Jiang, M, Koga, R, Xiao, R, Ciccosanti, C, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-07-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Principles for designing ideal protein structures.
Nature, 491, 2012
|
|
4DQ1
 
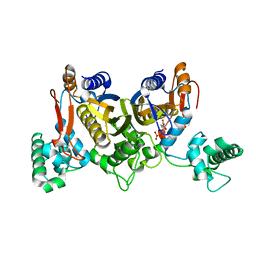 | | Thymidylate synthase from Staphylococcus aureus. | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Thymidylate synthase | | Authors: | Osipiuk, J, Holowicki, J, Jedrzejczak, R, Rubin, E, Guinn, K, Ioerger, T, Baker, D, Sacchettini, J, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Structures of Mtb Proteins Conferring Susceptibility to Known Mtb Inhibitors (MTBI) | | Deposit date: | 2012-02-14 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Thymidylate synthase from Staphylococcus aureus.
To be Published
|
|
2WPT
 
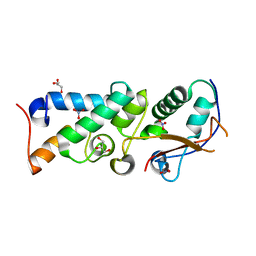 | | The crystal structure of Im2 in complex with colicin E9 DNase | | Descriptor: | COLICIN-E2 IMMUNITY PROTEIN, COLICIN-E9, GLYCEROL, ... | | Authors: | Meenan, N.A, Sharma, A, Fleishman, S.J, Macdonald, C.J, Boetzel, R, Moore, G.R, Baker, D, Kleanthous, C. | | Deposit date: | 2009-08-10 | | Release date: | 2010-06-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Structural and Energetic Basis for High Selectivity in a High-Affinity Protein-Protein Interaction.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4EGG
 
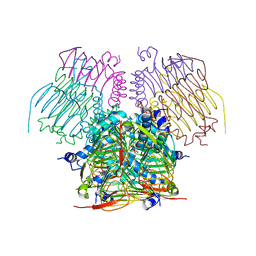 | | Computationally Designed Self-assembling tetrahedron protein, T310 | | Descriptor: | GLYCEROL, Putative acetyltransferase SACOL2570 | | Authors: | Sawaya, M.R, King, N.P, Sheffler, W, Baker, D, Yeates, T.O. | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Computational design of self-assembling protein nanomaterials with atomic level accuracy.
Science, 336, 2012
|
|
6ZV9
 
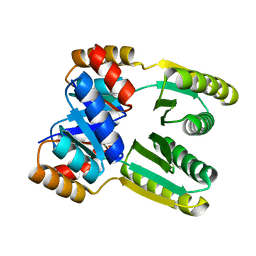 | | Terbium(III)-bound de novo TIM barrel-ferredoxin fold fusion dimer with 4-glutamate binding site and tryptophan antenna (TFD-EE N6W) | | Descriptor: | 1,2-ETHANEDIOL, TERBIUM(III) ION, TFD-EE | | Authors: | Caldwell, S, Haydon, I, Piperidou, N, Huang, P, Hilvert, D, Baker, D, Zeymer, C. | | Deposit date: | 2020-07-24 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Tight and specific lanthanide binding in a de novo TIM barrel with a large internal cavity designed by symmetric domain fusion.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6V67
 
 | | Apo Structure of the De Novo PD-1 Binding Miniprotein GR918.2 | | Descriptor: | PD-1 Binding Miniprotein GR918.2 | | Authors: | Bick, M.J, Bryan, C.M, Baker, D, Dimaio, F, Kang, A. | | Deposit date: | 2019-12-04 | | Release date: | 2020-12-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Computational design of a synthetic PD-1 agonist.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3J1W
 
 | |
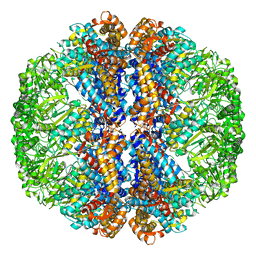
3J3X
 
 | |
8FVT
 
 | |
8GAA
 
 | |
8FG6
 
 | |
3SXW
 
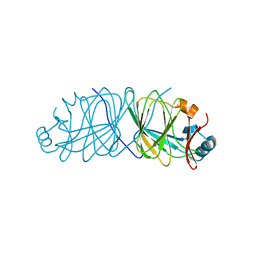 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR69. | | Descriptor: | Glucose-6-phosphate isomerase | | Authors: | Vorobiev, S, Su, M, Nivon, L, Seetharaman, J, Patel, P, Xiao, R, Maglaqui, M, Ciccosanti, C, Baker, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-07-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR69.
To be Published
|
|
8FWD
 
 | |
3TC6
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR63. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
3TC7
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR62. | | Descriptor: | ACETIC ACID, Indole-3-glycerol phosphate synthase, PHOSPHATE ION | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-08-08 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exploration of alternate catalytic mechanisms and optimization strategies for retroaldolase design.
J.Mol.Biol., 426, 2014
|
|
8FRE
 
 | | Designed loop protein RBL4 | | Descriptor: | 1,2-ETHANEDIOL, RBL4 | | Authors: | Jude, K.M, Jiang, H, Baker, D, Garcia, K.C. | | Deposit date: | 2023-01-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | De novo design of buttressed loops for sculpting protein functions.
Nat.Chem.Biol., 20, 2024
|
|
8FV5
 
 | | Representation of 16-mer phiPA3 PhuN Lattice, p2 | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein, phiPA3 PhuN | | Authors: | Nieweglowska, E.S, Brilot, A.F, Mendez-Moran, M, Kokontis, C, Baek, M, Li, J, Cheng, Y, Baker, D, Bondy-Denomy, J, Agard, D.A. | | Deposit date: | 2023-01-18 | | Release date: | 2023-03-01 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.21 Å) | | Cite: | The phi PA3 phage nucleus is enclosed by a self-assembling 2D crystalline lattice.
Nat Commun, 14, 2023
|
|
8FRF
 
 | | Homodimeric designed loop protein RBL7_C2_3 | | Descriptor: | MALONATE ION, RBL7_C2_3 | | Authors: | Jude, K.M, Jiang, H, Baker, D, Garcia, K.C. | | Deposit date: | 2023-01-06 | | Release date: | 2024-04-17 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | De novo design of buttressed loops for sculpting protein functions.
Nat.Chem.Biol., 20, 2024
|
|
4TQL
 
 | | Computationally designed three helix bundle | | Descriptor: | Three helix bundle | | Authors: | Nannenga, B.L, Oberdorfer, G, DiMaio, F, Baker, D, Gonen, T. | | Deposit date: | 2014-06-11 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High thermodynamic stability of parametrically designed helical bundles.
Science, 346, 2014
|
|
1HZ5
 
 | | CRYSTAL STRUCTURES OF THE B1 DOMAIN OF PROTEIN L FROM PEPTOSTREPTOCOCCUS MAGNUS, WITH A TYROSINE TO TRYPTOPHAN SUBSTITUTION | | Descriptor: | PROTEIN L, ZINC ION | | Authors: | O'Neill, J.W, Kim, D.E, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-01-23 | | Release date: | 2001-04-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the B1 domain of protein L from Peptostreptococcus magnus with a tyrosine to tryptophan substitution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1HZ6
 
 | | CRYSTAL STRUCTURES OF THE B1 DOMAIN OF PROTEIN L FROM PEPTOSTREPTOCOCCUS MAGNUS WITH A TYROSINE TO TRYPTOPHAN SUBSTITUTION | | Descriptor: | PROTEIN L | | Authors: | O'Neill, J.W, Kim, D.E, Baker, D, Zhang, K.Y.J. | | Deposit date: | 2001-01-23 | | Release date: | 2001-04-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the B1 domain of protein L from Peptostreptococcus magnus with a tyrosine to tryptophan substitution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
4S0J
 
 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: 11BIF, 42F, 79S, and 123V mutant | | Descriptor: | Threonine--tRNA ligase | | Authors: | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2014-12-31 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
4S03
 
 | | Biphenylalanine modified threonyl-tRNA synthetase from Pyrococcus abyssi: I11BIF, Y79I, and F123A mutant | | Descriptor: | Threonine--tRNA ligase | | Authors: | Pearson, A.D, Mills, J.H, Song, Y, Nasertorabi, F, Han, G.W, Baker, D, Stevens, R.C, Schultz, P.G. | | Deposit date: | 2014-12-30 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Transition states. Trapping a transition state in a computationally designed protein bottle.
Science, 347, 2015
|
|
