4V7G
 
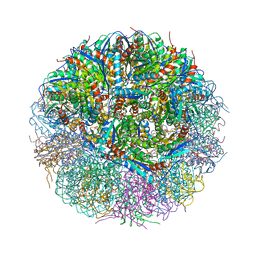 | | Crystal Structure of Lumazine Synthase from Bacillus Anthracis | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Morgunova, E, Illarionov, B, Saller, S, Popov, A, Sambaiah, T, Bacher, A, Cushman, M, Fischer, M, Ladenstein, R. | | Deposit date: | 2009-09-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural study and thermodynamic characterization of inhibitor binding to lumazine synthase from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
1A9C
 
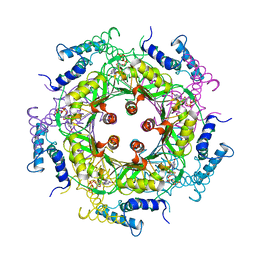 | | GTP CYCLOHYDROLASE I (C110S MUTANT) IN COMPLEX WITH GTP | | Descriptor: | GTP CYCLOHYDROLASE I, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Auerbach, G, Nar, H, Bracher, A, Bacher, A, Huber, R. | | Deposit date: | 1998-04-04 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | GTP Cyclohydrolase I in Complex with GTP at 2.1 A Resolution
To be Published
|
|
1A8R
 
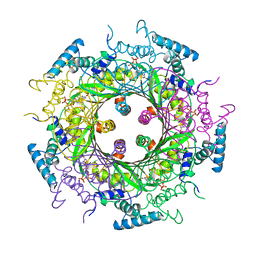 | | GTP CYCLOHYDROLASE I (H112S MUTANT) IN COMPLEX WITH GTP | | Descriptor: | GTP CYCLOHYDROLASE I, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Auerbach, G, Nar, H, Bracher, A, Bacher, A, Huber, R. | | Deposit date: | 1998-03-27 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biosynthesis of pteridines. Reaction mechanism of GTP cyclohydrolase I.
J.Mol.Biol., 326, 2003
|
|
4MUY
 
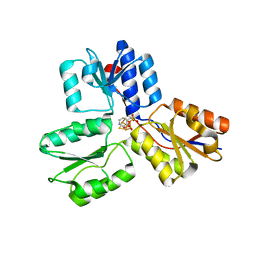 | | IspH in complex with pyridin-4-ylmethyl diphosphate | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER, pyridin-4-ylmethyl trihydrogen diphosphate | | Authors: | Span, I, Wang, K, Song, Y, Eisenreich, W, Bacher, A, Oldfield, E, Groll, M. | | Deposit date: | 2013-09-23 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into the Binding of Pyridines to the Iron-Sulfur Enzyme IspH.
J.Am.Chem.Soc., 136, 2014
|
|
4MV0
 
 | | IspH in complex with pyridin-2-ylmethyl diphosphate | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER, pyridin-2-ylmethyl trihydrogen diphosphate | | Authors: | Span, I, Wang, K, Song, Y, Eisenreich, W, Bacher, A, Oldfield, E, Groll, M. | | Deposit date: | 2013-09-23 | | Release date: | 2014-06-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the Binding of Pyridines to the Iron-Sulfur Enzyme IspH.
J.Am.Chem.Soc., 136, 2014
|
|
4MV5
 
 | | IspH in complex with 6-chloropyridin-3-ylmethyl diphosphate | | Descriptor: | (6-chloropyridin-3-yl)methyl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | Authors: | Span, I, Wang, K, Song, Y, Eisenreich, W, Bacher, A, Oldfield, E, Groll, M. | | Deposit date: | 2013-09-23 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the Binding of Pyridines to the Iron-Sulfur Enzyme IspH.
J.Am.Chem.Soc., 136, 2014
|
|
2A58
 
 | | Structure of 6,7-Dimethyl-8-ribityllumazine synthase from Schizosaccharomyces pombe mutant W27Y with bound riboflavin | | Descriptor: | 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION, RIBOFLAVIN | | Authors: | Koch, M, Breithaupt, C, Gerhardt, S, Haase, I, Weber, S, Cushman, M, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase
Eur.J.Biochem., 271, 2004
|
|
2A57
 
 | | Structure of 6,7-Dimthyl-8-ribityllumazine synthase from Schizosaccharomyces pombe mutant W27Y with bound ligand 6-carboxyethyl-7-oxo-8-ribityllumazine | | Descriptor: | 3-[8-((2S,3S,4R)-2,3,4,5-TETRAHYDROXYPENTYL)-2,4,7-TRIOXO-1,3,8-TRIHYDROPTERIDIN-6-YL]PROPANOIC ACID, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Koch, M, Breithaupt, C, Gerhardt, S, Haase, I, Weber, S, Cushman, M, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase
Eur.J.Biochem., 271, 2004
|
|
2A59
 
 | | Structure of 6,7-Dimethyl-8-ribityllumazine synthase from Schizosaccharomyces pombe mutant W27Y with bound ligand 5-nitroso-6-ribitylamino-2,4(1H,3H)-pyrimidinedione | | Descriptor: | 5-NITROSO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase, PHOSPHATE ION | | Authors: | Koch, M, Breithaupt, C, Gerhardt, S, Haase, I, Weber, S, Cushman, M, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of charge transfer complex formation by riboflavin bound to 6,7-dimethyl-8-ribityllumazine synthase
Eur.J.Biochem., 271, 2004
|
|
3KEF
 
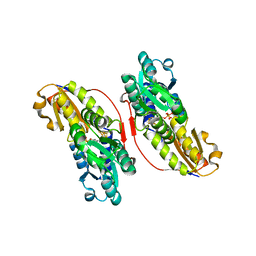 | | Crystal structure of IspH:DMAPP-complex | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, DIMETHYLALLYL DIPHOSPHATE, FE3-S4 CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KEM
 
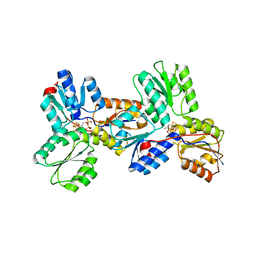 | | Crystal structure of IspH:IPP complex | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KE9
 
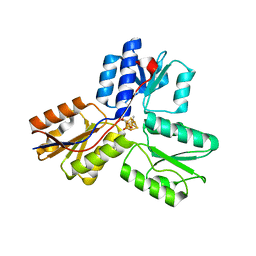 | | Crystal structure of IspH:Intermediate-complex | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-24 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KEL
 
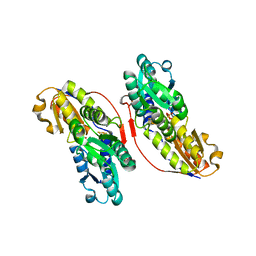 | | Crystal Structure of IspH:PP complex | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER, PYROPHOSPHATE 2- | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-26 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3KE8
 
 | | Crystal structure of IspH:HMBPP-complex | | Descriptor: | 4-HYDROXY-3-METHYL BUTYL DIPHOSPHATE, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Groll, M, Graewert, T, Span, I, Eisenreich, W, Bacher, A. | | Deposit date: | 2009-10-24 | | Release date: | 2010-01-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing the reaction mechanism of IspH protein by x-ray structure analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3DDY
 
 | | Structure of lumazine protein, an optical transponder of luminescent bacteria | | Descriptor: | Lumazine protein, RIBOFLAVIN | | Authors: | Chatwell, L, Illarionova, V, Illarionov, B, Skerra, A, Bacher, A, Fischer, M. | | Deposit date: | 2008-06-07 | | Release date: | 2008-07-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of lumazine protein, an optical transponder of luminescent bacteria.
J.Mol.Biol., 382, 2008
|
|
3R0I
 
 | | IspC in complex with an N-methyl-substituted hydroxamic acid | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, MANGANESE (II) ION, {(1S)-1-(3,4-difluorophenyl)-4-[hydroxy(methyl)amino]-4-oxobutyl}phosphonic acid | | Authors: | Behrendt, C.T, Kunfermann, A, Illarionova, V, Matheeussen, A, Pein, M.K, Graewert, T, Bacher, A, Eisenreich, W, Illarionov, B, Fischer, M, Maes, L, Groll, M, Kurz, T. | | Deposit date: | 2011-03-08 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Reverse Fosmidomycin Derivatives against the Antimalarial Drug Target IspC (Dxr).
J.Med.Chem., 54, 2011
|
|
3F7T
 
 | | Structure of active IspH shows a novel fold with a [3Fe-4S] cluster in the catalytic centre | | Descriptor: | 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, FE3-S4 CLUSTER, PHOSPHATE ION, ... | | Authors: | Graewert, T, Eppinger, J, Rohdich, F, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2008-11-10 | | Release date: | 2009-07-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of active IspH enzyme from Escherichia coli provides mechanistic insights into substrate reduction.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
3NOY
 
 | | Crystal structure of IspG (gcpE) | | Descriptor: | 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase, IRON/SULFUR CLUSTER | | Authors: | Groll, M, Graewert, T, Bacher, A. | | Deposit date: | 2010-06-26 | | Release date: | 2010-11-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biosynthesis of isoprenoids: crystal structure of the [4Fe-4S] cluster protein IspG.
J.Mol.Biol., 404, 2010
|
|
1SNN
 
 | | 3,4-dihydroxy-2-butanone 4-phosphate synthase from Methanococcus jannaschii | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, CALCIUM ION, RIBULOSE-5-PHOSPHATE, ... | | Authors: | Steinbacher, S, Schiffmann, S, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2004-03-11 | | Release date: | 2004-07-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Metal sites in 3,4-dihydroxy-2-butanone 4-phosphate synthase from Methanococcus jannaschii in complex with the substrate ribulose 5-phosphate.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5MRQ
 
 | | Arabidopsis thaliana IspD Asp262Ala Mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, ... | | Authors: | Schwab, A, Illarionov, B, Frank, A, Kunfermann, A, Seet, M, Bacher, A, Witschel, M, Fischer, M, Groll, M, Diederich, F. | | Deposit date: | 2016-12-23 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Allosteric Inhibition of the Enzyme IspD by Three Different Classes of Ligands.
ACS Chem. Biol., 12, 2017
|
|
5MRP
 
 | | Arabidopsis thaliana IspD Glu258Ala mutant in complex with Azolopyrimidine (2) | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, 5-chloro-7-hydroxy-6-(phenylmethyl)pyrazolo[1,5-a]pyrimidine-3-carbonitrile, ... | | Authors: | Schwab, A, Illarionov, B, Frank, A, Kunfermann, A, Seet, M, Bacher, A, Witschel, M, Fischer, M, Groll, M, Diederich, F. | | Deposit date: | 2016-12-23 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of Allosteric Inhibition of the Enzyme IspD by Three Different Classes of Ligands.
ACS Chem. Biol., 12, 2017
|
|
5MRM
 
 | | Arabidopsis thaliana IspD in complex with Isoxazole (4) | | Descriptor: | 2,4-bis(bromanyl)-6-[3-(trifluoromethyl)-1,2-oxazol-5-yl]phenol, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, ... | | Authors: | Schwab, A, Illarionov, B, Frank, A, Kunfermann, A, Seet, M, Bacher, A, Witschel, M, Fischer, M, Groll, M, Diederich, F. | | Deposit date: | 2016-12-23 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Allosteric Inhibition of the Enzyme IspD by Three Different Classes of Ligands.
ACS Chem. Biol., 12, 2017
|
|
5MRO
 
 | | Arabidopsis thaliana IspD Glu258Ala mutant in complex with Azolopyrimidine (1) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, ... | | Authors: | Schwab, A, Illarionov, B, Frank, A, Kunfermann, A, Seet, M, Bacher, A, Witschel, M, Fischer, M, Groll, M, Diederich, F. | | Deposit date: | 2016-12-23 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of Allosteric Inhibition of the Enzyme IspD by Three Different Classes of Ligands.
ACS Chem. Biol., 12, 2017
|
|
5MRN
 
 | | Arabidopsis thaliana IspD Glu258Ala Mutant | | Descriptor: | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase, chloroplastic, CADMIUM ION, ... | | Authors: | Schwab, A, Illarionov, B, Frank, A, Kunfermann, A, Seet, M, Bacher, A, Witschel, M, Fischer, M, Groll, M, Diederich, F. | | Deposit date: | 2016-12-23 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanism of Allosteric Inhibition of the Enzyme IspD by Three Different Classes of Ligands.
ACS Chem. Biol., 12, 2017
|
|
1EJB
 
 | | LUMAZINE SYNTHASE FROM SACCHAROMYCES CEREVISIAE | | Descriptor: | 5-(6-D-RIBITYLAMINO-2,4-DIHYDROXYPYRIMIDIN-5-YL)-1-PENTYL-PHOSPHONIC ACID, LUMAZINE SYNTHASE | | Authors: | Meining, W, Mortl, S, Fischer, M, Cushman, M, Bacher, A, Ladenstein, R. | | Deposit date: | 2000-03-02 | | Release date: | 2001-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The atomic structure of pentameric lumazine synthase from Saccharomyces cerevisiae at 1.85 A resolution reveals the binding mode of a phosphonate intermediate analogue.
J.Mol.Biol., 299, 2000
|
|
