4URI
 
 | | Crystal structure of chitinase-like agglutinin RobpsCRA from Robinia pseudoacacia | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHITINASE-RELATED AGGLUTININ, CHLORIDE ION, ... | | Authors: | Sulzenbacher, G, Roig-Zamboni, V, Peumans, W.J, Henrissat, B, van Damme, E.J.M, Bourne, Y. | | Deposit date: | 2014-06-30 | | Release date: | 2015-03-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis for Carbohydrate Binding Properties of a Plant Chitinase-Like Agglutinin with Conserved Catalytic Machinery.
J.Struct.Biol., 190, 2015
|
|
4V3B
 
 | | The structure of alpha2,3-sialyltransferase variant 1 from Pasteurella dagmatis in complex with the donor product CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
4V31
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Deoxyuridine | | Descriptor: | 2'-DEOXYURIDINE, CEREBLON ISOFORM 4, CITRATE ANION, ... | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2014-10-15 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Thalidomide Mimics Uridine Binding to an Aromatic Cage in Cereblon.
J.Struct.Biol., 188, 2014
|
|
4V2Z
 
 | |
4V32
 
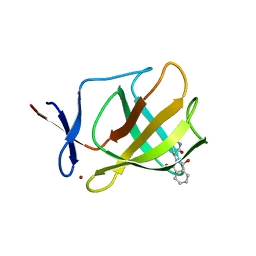 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Thalidomide, Y101F mutant | | Descriptor: | CEREBLON ISOFORM 4, S-Thalidomide, ZINC ION | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2014-10-15 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thalidomide Mimics Uridine Binding to an Aromatic Cage in Cereblon.
J.Struct.Biol., 188, 2014
|
|
4WFS
 
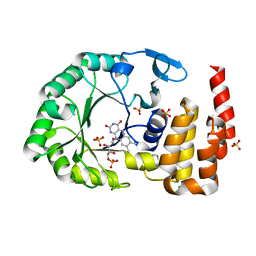 | | Crystal Structure of tRNA-dihydrouridine(20) synthase catalytic domain | | Descriptor: | CHLORIDE ION, FLAVIN MONONUCLEOTIDE, SULFATE ION, ... | | Authors: | Bou-Nader, C, Pecqueur, L, Kamah, A, Bregeon, D, Golinelli-Pimpaneau, B, Guimaraes, B.G, Fontecave, M, Hamdane, D. | | Deposit date: | 2014-09-17 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | An extended dsRBD is required for post-transcriptional modification in human tRNAs.
Nucleic Acids Res., 43, 2015
|
|
4WG7
 
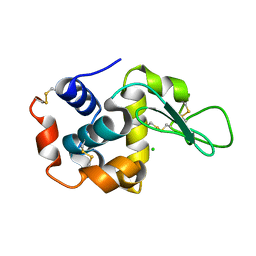 | | Room-temperature crystal structure of lysozyme determined by serial synchrotron crystallography using a nano focused beam. | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Coquelle, N, Brewster, A.S, Kappe, U, Shilova, A, Weinhausen, B, Burghammer, M, Colletier, J.P. | | Deposit date: | 2014-09-18 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Raster-scanning serial protein crystallography using micro- and nano-focused synchrotron beams.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WJ5
 
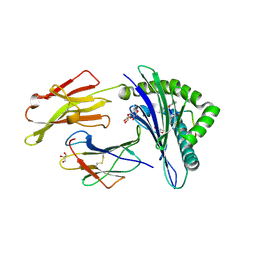 | | Structure of HLA-A2 in complex with an altered peptide ligands based on Mart-1 variant epitope | | Descriptor: | Beta-2-microglobulin, GLYCEROL, HLA class I histocompatibility antigen, ... | | Authors: | Celie, P.H.N, Rodenko, B, Ovaa, H. | | Deposit date: | 2014-09-29 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Altered Peptide Ligands Revisited: Vaccine Design through Chemically Modified HLA-A2-Restricted T Cell Epitopes.
J Immunol., 193, 2014
|
|
4WFD
 
 | | Structure of the Rrp6-Rrp47-Mtr4 interaction | | Descriptor: | ATP-dependent RNA helicase DOB1, Exosome complex exonuclease RRP6, Exosome complex protein LRP1, ... | | Authors: | Schuch, B, Conti, E. | | Deposit date: | 2014-09-14 | | Release date: | 2014-10-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase.
Embo J., 33, 2014
|
|
4WFT
 
 | | Crystal structure of tRNA-dihydrouridine(20) synthase dsRBD domain | | Descriptor: | tRNA-dihydrouridine(20) synthase [NAD(P)+]-like | | Authors: | Bou-Nader, C, Pecqueur, L, Kamah, A, Bregeon, D, Golinelli-Pimpaneau, B, Guimaraes, B.G, Fontecave, M, Hamdane, D. | | Deposit date: | 2014-09-17 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An extended dsRBD is required for post-transcriptional modification in human tRNAs.
Nucleic Acids Res., 43, 2015
|
|
4WL6
 
 | | Raster-scanning protein crystallography using micro and nano-focused synchrotron beams | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Coquelle, N, Kapp, U, Shilova, A, Weinhausen, B, Burghammer, M, Colletier, J.P. | | Deposit date: | 2014-10-06 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Raster-scanning serial protein crystallography using micro- and nano-focused synchrotron beams.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WZ2
 
 | | Crystal structure of U-box 2 of LubX / LegU2 / Lpp2887 from Legionella pneumophila str. Paris, Ile175Met mutant | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase LubX, HEXANE-1,6-DIOL | | Authors: | Stogios, P.J, Qualie, A.T, Skarina, T, Nocek, B, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-18 | | Release date: | 2015-01-28 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
4X4X
 
 | |
4WFC
 
 | | Structure of the Rrp6-Rrp47 interaction | | Descriptor: | Exosome complex exonuclease RRP6, Exosome complex protein LRP1, SULFATE ION | | Authors: | Schuch, B, Conti, E. | | Deposit date: | 2014-09-14 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The exosome-binding factors Rrp6 and Rrp47 form a composite surface for recruiting the Mtr4 helicase.
Embo J., 33, 2014
|
|
4WL7
 
 | | Room-temperature crystal structure of lysozyme determined by serial synchrotron crystallography using a micro focused beam (Conventional resolution cut-off) | | Descriptor: | CHLORIDE ION, Lysozyme C | | Authors: | Coquelle, N, Brewster, A.S, Kapp, U, Shilova, A, Weinhausen, B, Burghammer, M, Colletier, J.P. | | Deposit date: | 2014-10-06 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Raster-scanning serial protein crystallography using micro- and nano-focused synchrotron beams.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WRD
 
 | |
4WZ3
 
 | | Crystal structure of the complex between LubX/LegU2/Lpp2887 U-box 1 and Homo sapiens UBE2D2 | | Descriptor: | E3 ubiquitin-protein ligase LubX, Ubiquitin-conjugating enzyme E2 D2 | | Authors: | Stogios, P.J, Quaile, A.T, Skarina, T, Nocek, B, Di Leo, R, Yim, V, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-11-18 | | Release date: | 2015-01-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular Characterization of LubX: Functional Divergence of the U-Box Fold by Legionella pneumophila.
Structure, 23, 2015
|
|
4X0W
 
 | | The crystal structure of mupain-1-17 in complex with murinised human uPA | | Descriptor: | SULFATE ION, Urokinase-type plasminogen activator, mupain-1-17, ... | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-24 | | Release date: | 2015-10-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinctive binding modes and inhibitory mechanisms of two peptidic inhibitors of urokinase-type plasminogen activator with isomeric P1 residues.
Int.J.Biochem.Cell Biol., 62, 2015
|
|
4X1P
 
 | | The crystal structure of mupain-1-17 in complex with murinised human uPA at pH4.6 | | Descriptor: | MUPAIN-1-17, SULFATE ION, TRIETHYLENE GLYCOL, ... | | Authors: | Jiang, L, Zhao, B, Xu, P, Andreasen, P, Huang, M. | | Deposit date: | 2014-11-25 | | Release date: | 2015-10-21 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Distinctive binding modes and inhibitory mechanisms of two peptidic inhibitors of urokinase-type plasminogen activator with isomeric P1 residues.
Int.J.Biochem.Cell Biol., 62, 2015
|
|
7ZPG
 
 | | CRYSTAL STRUCTURE OF HUMAN MONOGLYCERIDE LIPASE WITH LIGAND | | Descriptor: | Monoglyceride lipase, [(7R,9aR)-7-(4-chlorophenyl)-1,3,4,6,7,8,9,9a-octahydropyrido[1,2-a]pyrazin-2-yl]-(2-bromanyl-3-methoxy-phenyl)methanone | | Authors: | Kemble, A, Hornsperger, B, Ruf, I, Richter, H, Benz, J, Kuhn, B, Heer, D, Wittwer, M, Engelhardt, B, Grether, U, Collin, L, Leibrock, L. | | Deposit date: | 2022-04-27 | | Release date: | 2022-09-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | A potent and selective inhibitor for the modulation of MAGL activity in the neurovasculature.
Plos One, 17, 2022
|
|
4WQ4
 
 | | E. coli YgjD(E12A)-YeaZ heterodimer in complex with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, W, Collinet, B. | | Deposit date: | 2014-10-21 | | Release date: | 2015-01-28 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The ATP-mediated formation of the YgjD-YeaZ-YjeE complex is required for the biosynthesis of tRNA t6A in Escherichia coli.
Nucleic Acids Res., 43, 2015
|
|
4X33
 
 | | Structure of the Elongator cofactor complex Kti11/Kti13 at 1.45A | | Descriptor: | 1,2-DIMETHOXYETHANE, CHLORIDE ION, Diphthamide biosynthesis protein 3, ... | | Authors: | Kolaj-Robin, O, McEwen, A.G, Cavarelli, J, Seraphin, B. | | Deposit date: | 2014-11-27 | | Release date: | 2015-01-21 | | Last modified: | 2015-03-11 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the Elongator cofactor complex Kti11/Kti13 provides insight into the role of Kti13 in Elongator-dependent tRNA modification.
Febs J., 282, 2015
|
|
4X57
 
 | |
4XE3
 
 | | OleP, the cytochrome P450 epoxidase from Streptomyces antibioticus involved in Oleandomycin biosynthesis: functional analysis and crystallographic structure in complex with clotrimazole. | | Descriptor: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, Cytochrome P-450, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Montemiglio, L.C, Parisi, G, Scaglione, A, Savino, C, Vallone, B. | | Deposit date: | 2014-12-22 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Functional analysis and crystallographic structure of clotrimazole bound OleP, a cytochrome P450 epoxidase from Streptomyces antibioticus involved in oleandomycin biosynthesis.
Biochim.Biophys.Acta, 1860, 2015
|
|
4XCU
 
 | | Crystal Structure of FGFR4 with an Irreversible Inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-(2-{[6-(2,6-dichloro-3,5-dimethoxyphenyl)quinazolin-2-yl]amino}-3-methylphenyl)propanamide, SULFATE ION | | Authors: | Kim, J.L, Miduturu, C, Hodous, B, Brooijmans, N, Bifulco, N, Guzi, T. | | Deposit date: | 2014-12-18 | | Release date: | 2015-04-01 | | Last modified: | 2018-12-12 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | First Selective Small Molecule Inhibitor of FGFR4 for the Treatment of Hepatocellular Carcinomas with an Activated FGFR4 Signaling Pathway.
Cancer Discov, 5, 2015
|
|
