3ESC
 
 | | cut-2a; NCN-Pt-Pincer-Cutinase Hybrid | | Descriptor: | Cutinase 1, bromo(4-{3-[(R)-ethoxy(4-nitrophenoxy)phosphoryl]propyl}-2,6-bis[(methylsulfanyl-kappaS)methyl]phenyl-kappaC~1~)palladium(2+) | | Authors: | Rutten, L, Mannie, J.P.B.A, Lutz, M, Gros, P. | | Deposit date: | 2008-10-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Solid-state structural characterization of cutinase-ECE-pincer-metal hybrids
Chemistry, 15, 2009
|
|
2WAB
 
 | | Structure of an active site mutant of a family two carbohydrate esterase from Clostridium thermocellum in complex with celluohexase | | Descriptor: | ENDOGLUCANASE E, GLYCEROL, IODIDE ION, ... | | Authors: | Montainer, C, Money, V.A, Pires, V.M.R, Flint, J.E, Pinheiro, B.A, Goyal, A, Prates, J.A.M, Izumi, A, Stalbrand, H, Kolenova, K, Topakas, E, Dodson, E.J, Bolam, D.N, Davies, G.J, Fontes, C.M.G.A, Gilbert, H.J. | | Deposit date: | 2009-02-04 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Active Site of a Carbohydrate Esterase Displays Divergent Catalytic and Noncatalytic Binding Functions.
Plos Biol., 7, 2009
|
|
3ESA
 
 | | cut-1b; NCN-Pt-Pincer-Cutinase Hybrid | | Descriptor: | (2,6-bis[(dimethylamino-kappaN)methyl]-4-{3-[(S)-ethoxy(4-nitrophenoxy)phosphoryl]propyl}phenyl-kappaC~1~)(chloro)platinum(2+), Cutinase 1 | | Authors: | Rutten, L, Mannie, J.P.B.A, Lutz, M, Gros, P. | | Deposit date: | 2008-10-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Solid-state structural characterization of cutinase-ECE-pincer-metal hybrids
Chemistry, 15, 2009
|
|
3ESB
 
 | | cut-1c; NCN-Pt-Pincer-Cutinase Hybrid | | Descriptor: | (2,6-bis[(dimethylamino-kappaN)methyl]-4-{3-[(S)-ethoxy(4-nitrophenoxy)phosphoryl]propyl}phenyl-kappaC~1~)(chloro)platinum(2+), CHLORIDE ION, Cutinase 1 | | Authors: | Rutten, L, Mannie, J.P.B.A, Lutz, M, Gros, P. | | Deposit date: | 2008-10-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Solid-state structural characterization of cutinase-ECE-pincer-metal hybrids
Chemistry, 15, 2009
|
|
5L9U
 
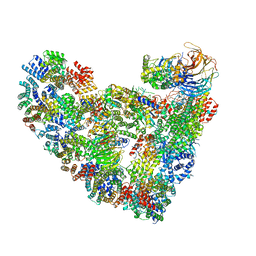 | | Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with a cross linked Ubiquitin variant-substrate-UBE2C (UBCH10) complex representing key features of multiubiquitination | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Brown, N.G, VanderLinden, R, Dube, P, Haselbach, D, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-09-14 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
5L9T
 
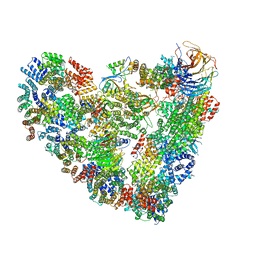 | | Model of human Anaphase-promoting complex/Cyclosome (APC/C-CDH1) with E2 UBE2S poised for polyubiquitination where UBE2S, APC2, and APC11 are modeled into low resolution density | | Descriptor: | Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Brown, N.G, VanderLinden, R, Dube, P, Haselbach, D, Peters, J.M, Stark, H, Schulman, B.A. | | Deposit date: | 2016-06-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual RING E3 Architectures Regulate Multiubiquitination and Ubiquitin Chain Elongation by APC/C.
Cell, 165, 2016
|
|
5M0Y
 
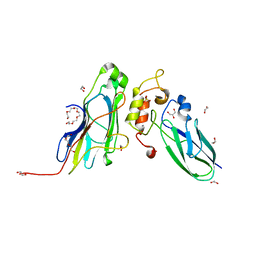 | | Crystal Structure of the CohScaA-XDocCipB type II complex from Clostridium thermocellum at 1.5Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cellulosome anchoring protein cohesin region, ... | | Authors: | Pinheiro, B.A, Bras, J.L, Carvalho, A.L, Fontes, C.M.G.A. | | Deposit date: | 2016-10-06 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Diverse specificity of cellulosome attachment to the bacterial cell surface.
Sci Rep, 6, 2016
|
|
5N16
 
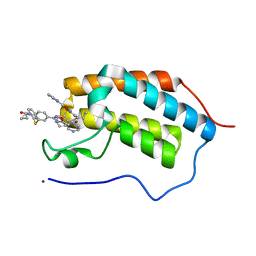 | | First Bromodomain (BD1) from Candida albicans Bdf1 bound to a dibenzothiazepinone (compound 1) | | Descriptor: | 5-cyclopropyl-2-(5-pyrazin-2-yl-1,2,4-oxadiazol-3-yl)benzo[b][1,4]benzothiazepin-6-one, Bromodomain-containing factor 1, GLYCEROL, ... | | Authors: | Mietton, F, Ferri, E, Champleboux, M, Zala, N, Maubon, D, Zhou, Y, Harbut, M, Spittler, D, Garnaud, C, Courcon, M, Chauvel, M, d'Enfert, C, Kashemirov, B.A, Hull, M, Cornet, M, McKenna, C.E, Govin, J, Petosa, C. | | Deposit date: | 2017-02-05 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Selective BET bromodomain inhibition as an antifungal therapeutic strategy.
Nat Commun, 8, 2017
|
|
7NS3
 
 | | Substrate receptor scaffolding module of yeast Chelator-GID SR4 E3 ubiquitin ligase bound to Fbp1 substrate | | Descriptor: | BJ4_G0018240.mRNA.1.CDS.1, Fructose-bisphosphatase, Glucose-induced degradation protein 8, ... | | Authors: | Sherpa, D, Chrustowicz, J, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-03-05 | | Release date: | 2021-05-05 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
7NSB
 
 | | Supramolecular assembly module of yeast Chelator-GID SR4 E3 ubiquitin ligase | | Descriptor: | Glucose-induced degradation protein 7, Glucose-induced degradation protein 8, Vacuolar import and degradation protein 30 | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-03-05 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
7NS5
 
 | | Structure of yeast Fbp1 (Fructose-1,6-bisphosphatase 1) | | Descriptor: | Fructose-1,6-bisphosphatase, MAGNESIUM ION, PHOSPHATE ION | | Authors: | Sherpa, D, Chrustowicz, J, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-03-05 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
7NSC
 
 | | Substrate receptor scaffolding module of human CTLH E3 ubiquitin ligase | | Descriptor: | Glucose-induced degradation protein 4 homolog, Glucose-induced degradation protein 8 homolog, Isoform 2 of Armadillo repeat-containing protein 8, ... | | Authors: | Chrustowicz, J, Sherpa, D, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-03-05 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
7NS4
 
 | | Catalytic module of yeast Chelator-GID SR4 E3 ubiquitin ligase | | Descriptor: | E3 ubiquitin-protein ligase RMD5, Protein FYV10, ZINC ION | | Authors: | Sherpa, D, Chrustowicz, J, Prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-03-05 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | GID E3 ligase supramolecular chelate assembly configures multipronged ubiquitin targeting of an oligomeric metabolic enzyme.
Mol.Cell, 81, 2021
|
|
2RMU
 
 | | THREE-DIMENSIONAL STRUCTURES OF DRUG-RESISTANT MUTANTS OF HUMAN RHINOVIRUS 14 | | Descriptor: | HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP1), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP2), HUMAN RHINOVIRUS 14 COAT PROTEIN (SUBUNIT VP3), ... | | Authors: | Badger, J, Krishnaswamy, S, Kremer, M.J, Oliveira, M.A, Rossmann, M.G, Heinz, B.A, Rueckert, R.R, Dutko, F.J, Mckinlay, M.A. | | Deposit date: | 1988-10-03 | | Release date: | 1990-01-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three-dimensional structures of drug-resistant mutants of human rhinovirus 14.
J.Mol.Biol., 207, 1989
|
|
4QX2
 
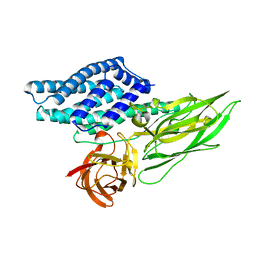 | | Cry3A Toxin structure obtained by injecting Bacillus thuringiensis cells in an XFEL beam, collecting data by serial femtosecond crystallographic methods and processing data with the cctbx.xfel software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4RF1
 
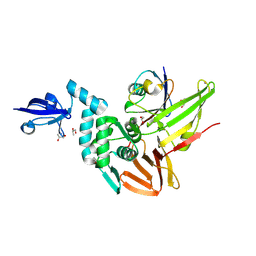 | | Crystal structure of the Middle-East respiratory syndrome coronavirus papain-like protease in complex with ubiquitin (space group P63) | | Descriptor: | 3-AMINOPROPANE, ORF1ab protein, S-1,2-PROPANEDIOL, ... | | Authors: | Bailey-Elkin, B.A, Johnson, G.G, Mark, B.L. | | Deposit date: | 2014-09-24 | | Release date: | 2014-10-22 | | Last modified: | 2015-01-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Papain-like Protease Bound to Ubiquitin Facilitates Targeted Disruption of Deubiquitinating Activity to Demonstrate Its Role in Innate Immune Suppression.
J.Biol.Chem., 289, 2014
|
|
8FEC
 
 | | Structure of J-PKAc chimera complexed with Aplithianine derivative | | Descriptor: | 6-[(6P)-6-(4-bromo-1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-7H-purine, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Du, L, Wilson, B.A.P, Li, N, Martinez Fiesco, J.A, Dalilian, M, Wang, D, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | Deposit date: | 2022-12-06 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery and Synthesis of a Naturally Derived Protein Kinase Inhibitor that Selectively Inhibits Distinct Classes of Serine/Threonine Kinases.
J.Nat.Prod., 86, 2023
|
|
8FE2
 
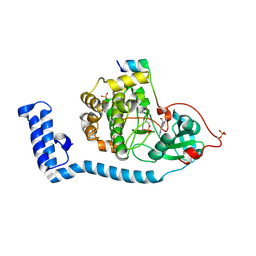 | | Structure of J-PKAc chimera complexed with Aplithianine A | | Descriptor: | 6-[(6M)-6-(1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-9H-purine, DnaJ homolog subfamily B member 1, cAMP-dependent protein kinase catalytic subunit alpha, ... | | Authors: | Du, L, Wilson, B.A.P, Li, N, Dalilian, M, Wang, D, Martinez Fiesco, J.A, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | Deposit date: | 2022-12-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Discovery and Synthesis of a Naturally Derived Protein Kinase Inhibitor that Selectively Inhibits Distinct Classes of Serine/Threonine Kinases.
J.Nat.Prod., 86, 2023
|
|
8FE5
 
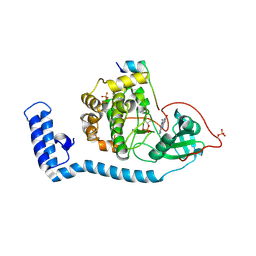 | | Structure of J-PKAc chimera complexed with Aplithianine B | | Descriptor: | 6-[(6P)-6-(1-methyl-1H-imidazol-5-yl)-2,3-dihydro-4H-1,4-thiazin-4-yl]-7,9-dihydro-8H-purin-8-one, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Du, L, Wilson, B.A.P, Li, N, Martinez Fiesco, J.A, Dalilian, M, Wang, D, Smith, E.A, Wamiru, A, Goncharova, E.I, Zhang, P, O'Keefe, B.R. | | Deposit date: | 2022-12-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Discovery and Synthesis of a Naturally Derived Protein Kinase Inhibitor that Selectively Inhibits Distinct Classes of Serine/Threonine Kinases.
J.Nat.Prod., 86, 2023
|
|
4QX1
 
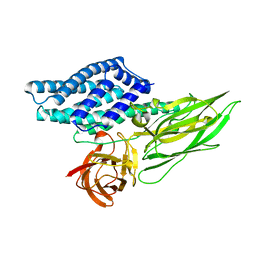 | | Cry3A Toxin structure obtained by Serial Femtosecond Crystallography from in vivo grown crystals isolated from Bacillus thuringiensis and data processed with the CrystFEL software suite | | Descriptor: | Pesticidal crystal protein cry3Aa | | Authors: | Sawaya, M.R, Cascio, D, Gingery, M, Rodriguez, J, Goldschmidt, L, Colletier, J.-P, Messerschmidt, M, Boutet, S, Koglin, J.E, Williams, G.J, Brewster, A.S, Nass, K, Hattne, J, Botha, S, Doak, R.B, Shoeman, R.L, DePonte, D.P, Park, H.-W, Federici, B.A, Sauter, N.K, Schlichting, I, Eisenberg, D. | | Deposit date: | 2014-07-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein crystal structure obtained at 2.9 angstrom resolution from injecting bacterial cells into an X-ray free-electron laser beam.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4REZ
 
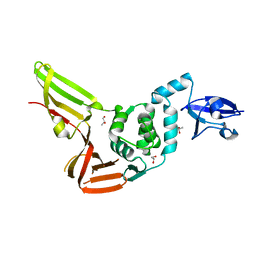 | | Crystal structure of the Middle-East respiratory syndrome coronavirus papain-like protease | | Descriptor: | ORF1ab protein, S-1,2-PROPANEDIOL, ZINC ION | | Authors: | Bailey-Elkin, B.A, Johnson, G.G, Mark, B.L. | | Deposit date: | 2014-09-24 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Papain-like Protease Bound to Ubiquitin Facilitates Targeted Disruption of Deubiquitinating Activity to Demonstrate Its Role in Innate Immune Suppression.
J.Biol.Chem., 289, 2014
|
|
8G32
 
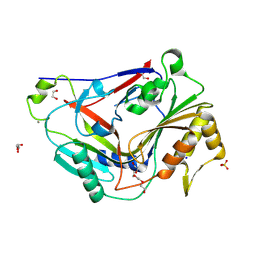 | | Pro-form of a CDCL short from E. anophelis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Johnstone, B.A, Christie, M.P, Morton, C.J, Parker, M.W. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Distant relatives of a eukaryotic cell-specific toxin family evolved a complement-like mechanism to kill bacteria.
Nat Commun, 15, 2024
|
|
8G33
 
 | |
4RG6
 
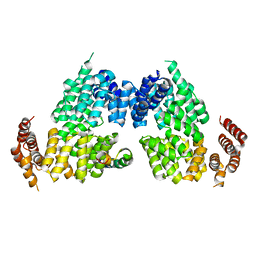 | | Crystal structure of APC3-APC16 complex | | Descriptor: | Anaphase-promoting complex subunit 16, Cell division cycle protein 27 homolog | | Authors: | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
4RG9
 
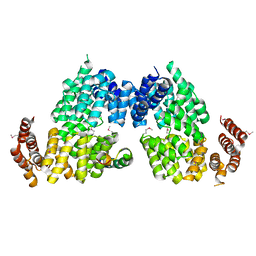 | | Crystal structure of APC3-APC16 complex (Selenomethionine Derivative) | | Descriptor: | Anaphase-promoting complex subunit 16, Cell division cycle protein 27 homolog | | Authors: | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
