7O8C
 
 | | Structure of SGBP BO2743 from Bacteroides ovatus | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, AZIDE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Correia, V.C, Trovao, F, Pinheiro, B.A, Palma, A.S, Carvalho, A.L. | | Deposit date: | 2021-04-15 | | Release date: | 2021-12-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping Molecular Recognition of beta 1,3-1,4-Glucans by a Surface Glycan-Binding Protein from the Human Gut Symbiont Bacteroides ovatus.
Microbiol Spectr, 9, 2021
|
|
5J54
 
 | | The Structure and Mechanism of NOV1, a Resveratrol-Cleaving Dioxygenase | | Descriptor: | Carotenoid oxygenase, FE (III) ION, OXYGEN MOLECULE, ... | | Authors: | McAndrew, R.P, Pereira, J.H, Sathitsuksanoh, N, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2016-04-01 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
1D8M
 
 | | CRYSTAL STRUCTURE OF MMP3 COMPLEXED WITH A HETEROCYCLE-BASED INHIBITOR | | Descriptor: | 1-BENZYL-3-(4-METHOXY-BENZENESULFONYL)-6-OXO-HEXAHYDRO-PYRIMIDINE-4-CARBOXYLIC ACID HYDROXYAMIDE, CALCIUM ION, STROMELYSIN-1 PRECURSOR, ... | | Authors: | Pikul, S, Dunham, K.M, Almstead, N.G, De, B, Natchus, M.G, Taiwo, Y.O, Williams, L.E, Hynd, B.A, Hsieh, L.C, Janusz, M.J, Gu, F, Mieling, G.E. | | Deposit date: | 1999-10-25 | | Release date: | 2000-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Heterocycle-based MMP inhibitors with P2' substituents.
Bioorg.Med.Chem.Lett., 11, 2001
|
|
1CLF
 
 | | CLOSTRIDIUM PASTEURIANUM FERREDOXIN | | Descriptor: | FERREDOXIN, IRON/SULFUR CLUSTER | | Authors: | Bertini, I, Donaire, A, Feinberg, B.A, Luchinat, C, Piccioli, M, Yuan, H. | | Deposit date: | 1995-06-21 | | Release date: | 1996-01-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the oxidized 2[4Fe-4S] ferredoxin from Clostridium pasteurianum.
Eur.J.Biochem., 232, 1995
|
|
5J55
 
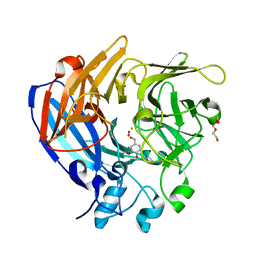 | | The Structure and Mechanism of NOV1, a Resveratrol-Cleaving Dioxygenase | | Descriptor: | 4-hydroxy-3-methoxybenzaldehyde, Carotenoid oxygenase, FE (III) ION, ... | | Authors: | McAndrew, R.P, Pereira, J.H, Sathitsuksanoh, N, Sale, K.L, Simmons, B.A, Adams, P.D. | | Deposit date: | 2016-04-01 | | Release date: | 2016-11-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and mechanism of NOV1, a resveratrol-cleaving dioxygenase.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
7ONI
 
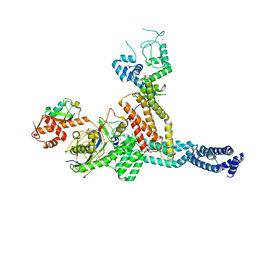 | | Structure of Neddylated CUL5 C-terminal region-RBX2-ARIH2* | | Descriptor: | Cullin-5, E3 ubiquitin-protein ligase ARIH2, NEDD8, ... | | Authors: | Kostrhon, S.P, prabu, J.R, Schulman, B.A. | | Deposit date: | 2021-05-25 | | Release date: | 2021-09-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | CUL5-ARIH2 E3-E3 ubiquitin ligase structure reveals cullin-specific NEDD8 activation.
Nat.Chem.Biol., 17, 2021
|
|
7WUG
 
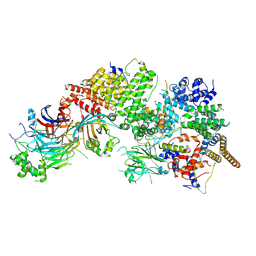 | | GID subcomplex: Gid12 bound Substrate Receptor Scaffolding module | | Descriptor: | Glucose-induced degradation protein 8, HLJ1_G0042170.mRNA.1.CDS.1, Vacuolar import and degradation protein 24, ... | | Authors: | Qiao, S, Cheng, J.D, Schulman, B.A. | | Deposit date: | 2022-02-08 | | Release date: | 2022-06-08 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structures of Gid12-bound GID E3 reveal steric blockade as a mechanism inhibiting substrate ubiquitylation.
Nat Commun, 13, 2022
|
|
6BG5
 
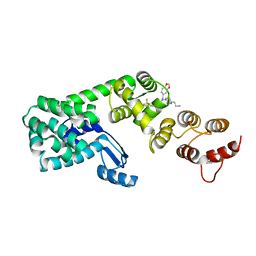 | | Structure of 1-(benzo[d][1,3]dioxol-5-ylmethyl)-1-(1-propylpiperidin-4-yl)-3-(3-(trifluoromethyl)phenyl)urea bound to DCN1 | | Descriptor: | Endolysin, DCN1-like protein 1 chimera, N-[(2H-1,3-benzodioxol-5-yl)methyl]-N-(1-propylpiperidin-4-yl)-N'-[3-(trifluoromethyl)phenyl]urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-10-27 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Piperidinyl Ureas Chemically Control Defective in Cullin Neddylation 1 (DCN1)-Mediated Cullin Neddylation.
J. Med. Chem., 61, 2018
|
|
6B8U
 
 | | Crystals Structure of B-Raf kinase domain in complex with an Imidazopyridinyl benzamide inhibitor | | Descriptor: | Serine/threonine-protein kinase B-raf, ~{N}-[3-(2-acetamidoimidazo[1,2-a]pyridin-6-yl)-4-methyl-phenyl]-3-(trifluoromethyl)benzamide | | Authors: | Appleton, B.A, Murray, J, Shafer, C.M. | | Deposit date: | 2017-10-09 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Imidazo[1,2-a]pyridin-6-yl-benzamide analogs as potent RAF inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7Z8B
 
 | |
8G32
 
 | | Pro-form of a CDCL short from E. anophelis | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Johnstone, B.A, Christie, M.P, Morton, C.J, Parker, M.W. | | Deposit date: | 2023-02-06 | | Release date: | 2024-02-07 | | Last modified: | 2024-06-26 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Distant relatives of a eukaryotic cell-specific toxin family evolved a complement-like mechanism to kill bacteria.
Nat Commun, 15, 2024
|
|
8G33
 
 | |
8GBR
 
 | |
4V3N
 
 | | Membrane bound pleurotolysin prepore (TMH2 strand lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V3A
 
 | | Membrane bound pleurotolysin prepore (TMH1 lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, CaradocDavies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-17 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
4V3M
 
 | | Membrane bound pleurotolysin prepore (TMH2 helix lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
8BD0
 
 | |
8BPI
 
 | |
7ODA
 
 | | OXA-48-like Beta-lactamase OXA-436 | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactamase, CHLORIDE ION | | Authors: | Lund, B.A, Thomassen, A.M, Carlsen, T.J.W, Leiros, H.K.S. | | Deposit date: | 2021-04-29 | | Release date: | 2021-09-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Biochemical and biophysical characterization of the OXA-48-like carbapenemase OXA-436.
Acta Crystallogr.,Sect.F, 77, 2021
|
|
7OD1
 
 | |
7O07
 
 | |
4LS9
 
 | | Structure of mycobacterial nrnA homolog reveals multifunctional nuclease activities | | Descriptor: | DHH family protein, GLYCEROL, MANGANESE (II) ION | | Authors: | Kumar, D, Srivastav, R, Grover, A, Manjasetty, B.A, Sharma, R, Taneja, B. | | Deposit date: | 2013-07-22 | | Release date: | 2014-07-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique subunit packing in mycobacterial nanoRNase leads to alternate substrate recognitions in DHH phosphodiesterases
Nucleic Acids Res., 42, 2014
|
|
4M18
 
 | | Crystal Structure of Surfactant Protein-D D325A/R343V mutant in complex with trimannose (Man-a1,2Man-a1,2Man) | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein D, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose, ... | | Authors: | Goh, B.C, Rynkiewicz, M.J, Cafarella, T.R, White, M.R, Hartshorn, K.L, Allen, K, Crouch, E.C, Calin, O, Seeberger, P.H, Schulten, K, Seaton, B.A. | | Deposit date: | 2013-08-02 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.203 Å) | | Cite: | Molecular mechanisms of inhibition of influenza by surfactant protein d revealed by large-scale molecular dynamics simulation.
Biochemistry, 52, 2013
|
|
7NOX
 
 | | Structure of SGBP BO2743 from Bacteroides ovatus in complex with mixed-linked gluco-nonasaccharide | | Descriptor: | AZIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Correia, V.C, Trovao, F, Pinheiro, B.A, Palma, A.S, Carvalho, A.L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Mapping Molecular Recognition of beta 1,3-1,4-Glucans by a Surface Glycan-Binding Protein from the Human Gut Symbiont Bacteroides ovatus.
Microbiol Spectr, 9, 2021
|
|
4V2T
 
 | | Membrane embedded pleurotolysin pore with 13 fold symmetry | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-15 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
