6TVQ
 
 | | Structure of native gp41 derived peptide fusion inhibitor | | Descriptor: | Env polyprotein (Fragment), Envelope glycoprotein gp160 | | Authors: | Huhmann, S, Nyakatura, E.K, Rohrhofer, A, Schmidt, B, Eichler, J, Moschner, J, Roth, C. | | Deposit date: | 2020-01-10 | | Release date: | 2021-01-27 | | Last modified: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Systematic Evaluation of Fluorination as Modification for Peptide-Based Fusion Inhibitors against HIV-1 Infection.
Chembiochem, 22, 2021
|
|
4MEV
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Rhodoferax ferrireducens (Rfer_1840), Target EFI-510211, with bound malonate, space group I422 | | Descriptor: | CITRIC ACID, MALONATE ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
3ZFJ
 
 | | N-terminal domain of pneumococcal PhtD protein with bound Zn(II) | | Descriptor: | PNEUMOCOCCAL HISTIDINE TRIAD PROTEIN D, ZINC ION | | Authors: | Bersch, B, Bougault, C, Favier, A, Gabel, F, Roux, L, Vernet, T, Durmort, C. | | Deposit date: | 2012-12-11 | | Release date: | 2013-11-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | New Insights Into Histidine Triad Proteins: Solution Structure of a Streptococcus Pneumoniae Phtd Domain and Zinc Transfer to Adcaii.
Plos One, 8, 2013
|
|
3ZCR
 
 | | Rabbit muscle glycogen phosphorylase b in complex with N-(4-tert- butyl-benzoyl)-N-beta-D-glucopyranosyl urea determined at 2.07 A resolution | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, INOSINIC ACID, ... | | Authors: | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2012-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
3ZCT
 
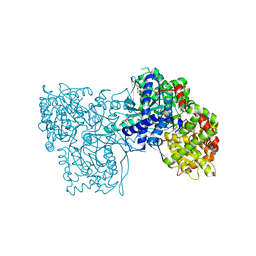 | | Rabbit muscle glycogen phosphorylase b in complex with N-(2-naphthoyl) -N-beta-D-glucopyranosyl urea determined at 2.0 A resolution | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, INOSINIC ACID, ... | | Authors: | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2012-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
3ZCQ
 
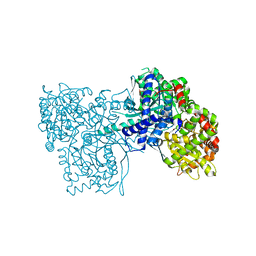 | | Rabbit muscle glycogen phosphorylase b in complex with N-(4- trifluoromethyl-benzoyl)-N-beta-D-glucopyranosyl urea determined at 2. 15 A resolution | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, N-{[4-(trifluoromethyl)benzoyl]carbamoyl}-beta-D-glucopyranosylamine, ... | | Authors: | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2012-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
2PRQ
 
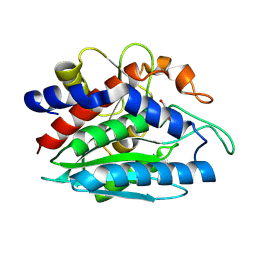 | | X-ray crystallographic characterization of the Co(II)-substituted Tris-bound form of the aminopeptidase from Aeromonas proteolytica | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bacterial leucyl aminopeptidase, COBALT (II) ION | | Authors: | Munih, P, Moulin, A, Stamper, C.C, Bennet, B, Ringe, D, Petsko, G.A, Holz, R.C. | | Deposit date: | 2007-05-04 | | Release date: | 2007-06-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | X-ray crystallographic characterization of the Co(II)-substituted Tris-bound form of the aminopeptidase from Aeromonas proteolytica.
J.Inorg.Biochem., 101, 2007
|
|
3ZKP
 
 | |
8CS8
 
 | | [High G:C, High Vapor Diffusion] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*AP*GP*CP*CP*TP*GP*CP*GP*CP*GP*GP*AP*CP*AP*GP*AP*G)-3'), DNA (5'-D(*TP*CP*CP*TP*CP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*CP*GP*CP*A)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-05-12 | | Release date: | 2022-12-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8CS4
 
 | | [AGC/GTC] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle | | Descriptor: | DNA (5'-D(*AP*GP*CP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*GP*TP*CP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-05-12 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.03 Å) | | Cite: | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8CYN
 
 | | [2T7+20bp Linker] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle with 20 bp Sticky-End Linker | | Descriptor: | DNA (5'-D(*GP*CP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-05-24 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (9.45 Å) | | Cite: | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
2PNM
 
 | |
8CS5
 
 | | [GGA/CTC] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle | | Descriptor: | DNA (5'-D(*CP*TP*CP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(*GP*GP*AP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-05-12 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (6.52 Å) | | Cite: | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8CS3
 
 | | [CAG/TCG] Self-Assembled 3D DNA Hexagonal Tensegrity Triangle | | Descriptor: | DNA (5'-D(*CP*AP*GP*CP*AP*GP*CP*CP*TP*GP*TP*AP*CP*GP*GP*AP*CP*AP*TP*CP*A)-3'), DNA (5'-D(*TP*CP*GP*TP*GP*AP*TP*GP*T)-3'), DNA (5'-D(P*CP*CP*GP*TP*AP*CP*A)-3'), ... | | Authors: | Lu, B, Vecchioni, S, Ohayon, Y.P, Seeman, N.C, Mao, C, Sha, R. | | Deposit date: | 2022-05-12 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (5.14 Å) | | Cite: | Programmable 3D Hexagonal Geometry of DNA Tensegrity Triangles.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
3ZCS
 
 | | Rabbit muscle glycogen phosphorylase b in complex with N-(1-naphthoyl) -N-beta-D-glucopyranosyl urea determined at 2.07 A resolution | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, INOSINIC ACID, ... | | Authors: | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2012-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
4MNC
 
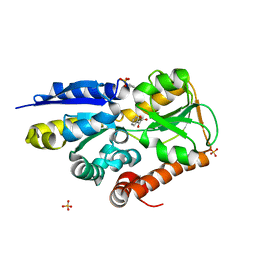 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_4736), Target EFI-510156, with bound benzoyl formate, space group P21 | | Descriptor: | BENZOYL-FORMIC ACID, SULFATE ION, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-10 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
3ZCV
 
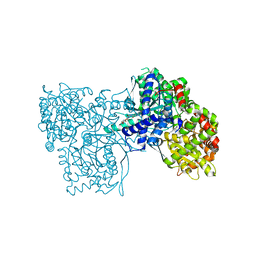 | | Rabbit muscle glycogen phosphorylase b in complex with N-(indol-2- carbonyl)-N-beta-D-glucopyranosyl urea determined at 1.8 A resolution | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, N-[(1H-indol-2-ylcarbonyl)carbamoyl]-beta-D-glucopyranosylamine, ... | | Authors: | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2012-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
4N4B
 
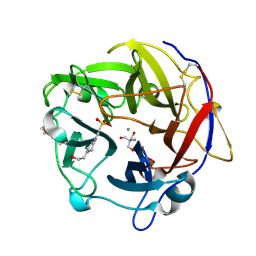 | | Crystal Structure of the alpha-L-arabinofuranosidase PaAbf62A from Podospora anserina | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Siguier, B, Dumon, C, Mourey, L, Tranier, S. | | Deposit date: | 2013-10-08 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | First Structural Insights into alpha-L-Arabinofuranosidases from the Two GH62 Glycoside Hydrolase Subfamilies.
J.Biol.Chem., 289, 2014
|
|
2PRN
 
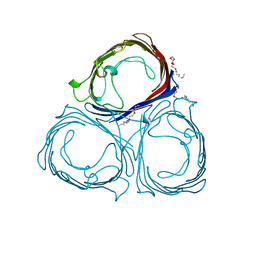 | | RHODOPSEUDOMONAS BLASTICA PORIN, TRIPLE MUTANT E1M, E99W, A116W | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, MAGNESIUM ION, PORIN | | Authors: | Maveyraud, L, Schmid, B, Schulz, G.E. | | Deposit date: | 1998-06-12 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Porin mutants with new channel properties.
Protein Sci., 7, 1998
|
|
4N15
 
 | | Crystal structure of a TRAP periplasmic solute binding protein from Burkholderia ambifaria (BAM_6123), Target EFI-510059, with bound beta-D-glucuronate | | Descriptor: | MAGNESIUM ION, TRAP dicarboxylate transporter, DctP subunit, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Zhao, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-03 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MIJ
 
 | | Crystal structure of a Trap periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_3107), target EFI-510173, with bound alpha/beta D-Galacturonate, space group P21 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, TRAP dicarboxylate transporter, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-08-31 | | Release date: | 2013-09-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4MNI
 
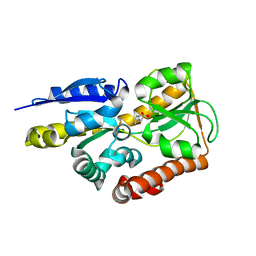 | | Crystal structure of a TRAP periplasmic solute binding protein from Polaromonas sp. JS666 (Bpro_4736), Target EFI-510156, with bound benzoyl formate, space group P6522 | | Descriptor: | BENZOYL-FORMIC ACID, TRAP dicarboxylate transporter-DctP subunit | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Zhao, S, Stead, M, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Jacobson, M.P, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-10 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
4N2Z
 
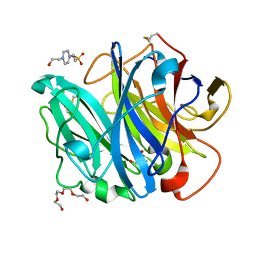 | | Crystal Structure of the alpha-L-arabinofuranosidase PaAbf62A from Podospora anserina in complex with cellotriose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Siguier, B, Dumon, C, Mourey, L, Tranier, S. | | Deposit date: | 2013-10-06 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | First Structural Insights into alpha-L-Arabinofuranosidases from the Two GH62 Glycoside Hydrolase Subfamilies.
J.Biol.Chem., 289, 2014
|
|
8DU3
 
 | | Crystal structure of A2AAR-StaR2-bRIL in complex with compound 21a | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (4M)-6-bromo-4-(furan-2-yl)quinazolin-2-amine, Adenosine receptor A2a, ... | | Authors: | Shiriaeva, A, Stauch, B, Han, G.W, Cherezov, V. | | Deposit date: | 2022-07-26 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High ligand efficiency quinazoline compounds as novel A 2A adenosine receptor antagonists.
Eur.J.Med.Chem., 241, 2022
|
|
8E1H
 
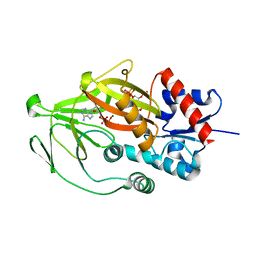 | | Asp1 kinase in complex with ADP Mg 5-IP7 | | Descriptor: | (1r,2R,3S,4s,5R,6S)-2,3,4,5,6-pentakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, ADENOSINE-5'-DIPHOSPHATE, Inositol hexakisphosphate and diphosphoinositol-pentakisphosphate kinase, ... | | Authors: | Goldgur, Y, Shuman, S, Benjamin, B. | | Deposit date: | 2022-08-10 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of Fission Yeast Inositol Pyrophosphate Kinase Asp1 in Ligand-Free, Substrate-Bound, and Product-Bound States.
Mbio, 13, 2022
|
|
