5A7Q
 
 | | Crystal structure of human JMJD2A in complex with compound 30 | | Descriptor: | 1,2-ETHANEDIOL, 2-(5-azanyl-2-oxidanyl-phenyl)pyridine-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Kopec, J, Dixon-Clarke, S, MacKenzie, A, Nowak, R, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | Deposit date: | 2015-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
3T9P
 
 | | Crystal structure of a putative mandelate racemase/muconate lactonizing enzyme family protein from Roseovarius | | Descriptor: | FORMIC ACID, GLYCEROL, Mandelate racemase/muconate lactonizing enzyme family protein, ... | | Authors: | Agarwal, R, Chamala, S, Evans, B, Foti, R, Gizzi, A, Hillerich, B, Kar, A, LaFleur, J, Seidel, R, Villigas, G, Zencheck, W, Almo, S.C, Swaminathan, S, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-08-03 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of a putative mandelate racemase/muconate lactonizing enzyme family protein from Roseovarius
To be Published
|
|
3TG0
 
 | | E. coli alkaline phosphatase with bound inorganic phosphate | | Descriptor: | Alkaline phosphatase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Bobyr, E, Lassila, J.K, Wiersma-Koch, H.I, Fenn, T.D, Lee, J.J, Nikolic-Hughes, I, Hodgson, K.O, Rees, D.C, Hedman, B, Herschlag, D. | | Deposit date: | 2011-08-16 | | Release date: | 2011-11-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution analysis of Zn(2+) coordination in the alkaline phosphatase superfamily by EXAFS and x-ray crystallography.
J.Mol.Biol., 415, 2012
|
|
2Y6K
 
 | | Xylotetraose bound to X-2 engineered mutated CBM4-2 Carbohydrate Binding Module from a Thermostable Rhodothermus marinus Xylanase | | Descriptor: | CALCIUM ION, CITRIC ACID, XYLANASE, ... | | Authors: | von Schantz, L, Hakansson, M, Logan, D.T, Walse, B, Osterlin, J, Nordberg-Karlsson, E, Ohlin, M. | | Deposit date: | 2011-01-24 | | Release date: | 2012-03-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural basis for carbohydrate-binding specificity--a comparative assessment of two engineered carbohydrate-binding modules.
Glycobiology, 22, 2012
|
|
4GQA
 
 | | Crystal structure of NAD binding oxidoreductase from Klebsiella pneumoniae | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NAD binding Oxidoreductase, ... | | Authors: | Osinski, S, Majorek, K.A, Niedzialkowska, E, Osinski, T, Porebski, P.J, Nawar, A, Hammonds, J, Hillerich, B, Seidel, R, Bonanno, J.B, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-08-22 | | Release date: | 2012-09-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of NAD binding oxidoreductase from Klebsiella pneumoniae (CASP Target)
To be Published
|
|
1G5N
 
 | | ANNEXIN V COMPLEX WITH HEPARIN OLIGOSACCHARIDES | | Descriptor: | 4-deoxy-2-O-sulfo-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ANNEXIN V, CALCIUM ION | | Authors: | Capila, I, Heraiz, M.J, Mo, Y.D, Mealy, T.R, Campos, B, Dedman, J.R, Linhardt, R.J, Seaton, B.A. | | Deposit date: | 2000-11-01 | | Release date: | 2001-06-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Annexin V--heparin oligosaccharide complex suggests heparan sulfate--mediated assembly on cell surfaces.
Structure, 9, 2001
|
|
1ONY
 
 | | Oxalyl-Aryl-Amino Benzoic Acid inhibitors of PTP1B, compound 17 | | Descriptor: | 2-{[2-(2-CARBAMOYL-VINYL)-4-(2-METHANESULFONYLAMINO-2-PENTYLCARBAMOYL-ETHYL)-PHENYL]-OXALYL-AMINO}-BENZOIC ACID, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Liu, G, Szczepankiewicz, B.G, Pei, Z, Janowich, D.A, Xin, Z, Hadjuk, P.J, Abad-Zapatero, C, Liang, H, Hutchins, C.W, Fesik, S.W, Ballaron, S.J, Stashko, M.A, Lubben, T, Mika, A.K, Zinker, B.A, Trevillyan, J.M, Jirousek, M.R. | | Deposit date: | 2003-03-02 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery and Structure-Activity Relationship of Oxalylarylaminobenzoic
Acids as Inhibitors of Protein Tyrosine Phosphatase 1B
J.Med.Chem., 46, 2003
|
|
3MEV
 
 | | Crystal structure of SGF29 in complex with R2AK4me3 | | Descriptor: | GLYCEROL, Histone H3, SAGA-associated factor 29 homolog, ... | | Authors: | Bian, C.B, Xu, C, Lam, R, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Sgf29 binds histone H3K4me2/3 and is required for SAGA complex recruitment and histone H3 acetylation.
Embo J., 30, 2011
|
|
6DYY
 
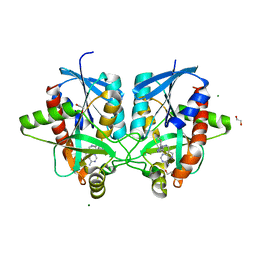 | | Crystal structure of Helicobacter pylori 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with (3R,4S)-1-((4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl)-4-(((3-(1-butyl-1H-1,2,3-triazol-4-yl)propyl)thio)methyl)pyrrolidin-3-ol | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-({[3-(1-butyl-1H-1,2,3-triazol-4-yl)propyl]sulfanyl}methyl)pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ... | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2018-07-02 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Selective Inhibitors of Helicobacter pylori Methylthioadenosine Nucleosidase and Human Methylthioadenosine Phosphorylase.
J. Med. Chem., 62, 2019
|
|
1GGZ
 
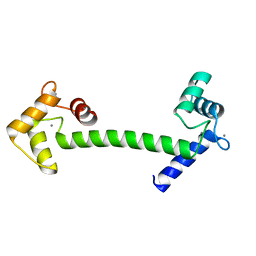 | | CRYSTAL STRUCTURE OF THE CALMODULIN-LIKE PROTEIN (HCLP) FROM HUMAN EPITHELIAL CELLS | | Descriptor: | CALCIUM ION, CALMODULIN-RELATED PROTEIN NB-1 | | Authors: | Han, B.-G, Han, M, Sui, H, Yaswen, P, Walian, P.J, Jap, B.K. | | Deposit date: | 2000-10-13 | | Release date: | 2002-06-05 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of human calmodulin-like protein: insights into its functional role.
FEBS Lett., 521, 2002
|
|
5IKR
 
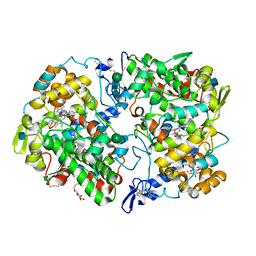 | | The Structure of Mefenamic Acid Bound to Human Cyclooxygenase-2 | | Descriptor: | 2-[(2,3-DIMETHYLPHENYL)AMINO]BENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, AMMONIUM ION, ... | | Authors: | Orlando, B.J, Malkowski, M.G. | | Deposit date: | 2016-03-03 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | Substrate-selective Inhibition of Cyclooxygeanse-2 by Fenamic Acid Derivatives Is Dependent on Peroxide Tone.
J.Biol.Chem., 291, 2016
|
|
1Y6Q
 
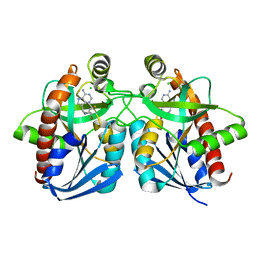 | | Cyrstal structure of MTA/AdoHcy nucleosidase complexed with MT-DADMe-ImmA | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, CHLORIDE ION, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Singh, V, Evans, G.B, Tyler, P.C, Furneaux, R.H, Cornell, K.A, Riscoe, M.K, Schramm, V.L, Howell, P.L. | | Deposit date: | 2004-12-06 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural rationale for the affinity of pico- and femtomolar transition state analogues of Escherichia coli 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase.
J.Biol.Chem., 280, 2005
|
|
1OGY
 
 | | Crystal structure of the heterodimeric nitrate reductase from Rhodobacter sphaeroides | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, DIHEME CYTOCHROME C NAPB MOLECULE: NITRATE REDUCTASE, HEME C, ... | | Authors: | Arnoux, P, Sabaty, M, Alric, J, Frangioni, B, Guigliarelli, B, Adriano, J.-M, Pignol, D. | | Deposit date: | 2003-05-19 | | Release date: | 2003-10-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural and Redox Plasticity in the Heterodimeric Periplasmic Nitrate Reductase
Nat.Struct.Biol., 10, 2003
|
|
5VLM
 
 | | Crystal structure of EilR in complex with crystal violet | | Descriptor: | CRYSTAL VIOLET, Regulatory protein TetR | | Authors: | Pereira, J.H, Ruegg, T.L, Chen, J, Novichkov, P, DeGiovani, A, Tomaleri, G.P, Singer, S, Simmons, B, Thelen, M, Adams, P.D. | | Deposit date: | 2017-04-25 | | Release date: | 2018-06-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.403 Å) | | Cite: | Jungle Express is a versatile repressor system for tight transcriptional control.
Nat Commun, 9, 2018
|
|
4EA3
 
 | | Structure of the N/OFQ Opioid Receptor in Complex with a Peptide Mimetic | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-benzyl-N-[3-(1'H,3H-spiro[2-benzofuran-1,4'-piperidin]-1'-yl)propyl]-D-prolinamide, ... | | Authors: | Thompson, A.A, Liu, W, Chun, E, Katritch, V, Wu, H, Vardy, E, Huang, X.P, Trapella, C, Guerrini, R, Calo, G, Roth, B.L, Cherezov, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2012-03-22 | | Release date: | 2012-04-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.013 Å) | | Cite: | Structure of the nociceptin/orphanin FQ receptor in complex with a peptide mimetic.
Nature, 485, 2012
|
|
2ZAD
 
 | |
5SX9
 
 | | Crystal Structure of PI3Kalpha in complex with fragment 14 | | Descriptor: | 4,6-dimethylpyridin-2-amine, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Gabelli, S.B, Vogelstein, B, Miller, M.S, Amzel, L.M. | | Deposit date: | 2016-08-09 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Identification of allosteric binding sites for PI3K alpha oncogenic mutant specific inhibitor design.
Bioorg. Med. Chem., 25, 2017
|
|
1GNG
 
 | | Glycogen synthase kinase-3 beta (GSK3) complex with FRATtide peptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FRATTIDE, GLYCOGEN SYNTHASE KINASE-3 BETA, ... | | Authors: | Bax, B, Carter, P.S, Lewis, C, Guy, A.R, Bridges, A, Tanner, R, Pettman, G, Mannix, C, Culbert, A.A, Brown, M.J.B, Smith, D.G, Reith, A.D. | | Deposit date: | 2001-10-04 | | Release date: | 2002-10-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure of Phosphorylated Gsk-3Beta Complexed with a Peptide, Frattide, that Inhibits Beta-Catenin Phosphorylation
Structure, 9, 2001
|
|
7KWY
 
 | | X-ray Crystal Structure of PlyCB Mutant R66K | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, PlyCB | | Authors: | Williams, D.E, Broendum, S.S, Hayes, B.K, Drinkwater, N, McGowan, S. | | Deposit date: | 2020-12-02 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High avidity drives the interaction between the streptococcal C1 phage endolysin, PlyC, with the cell surface carbohydrates of Group A Streptococcus.
Mol.Microbiol., 116, 2021
|
|
4BAG
 
 | |
6HX3
 
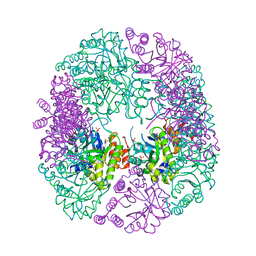 | | PDX1.2/PDX1.3 complex | | Descriptor: | Pyridoxal 5'-phosphate synthase subunit PDX1.3, Pyridoxal 5'-phosphate synthase-like subunit PDX1.2, SULFATE ION | | Authors: | Robinson, G.C, Kaufmann, M, Roux, C, Martinez-Font, J, Hothorn, M, Thore, S, Fitzpatrick, T.B. | | Deposit date: | 2018-10-15 | | Release date: | 2019-04-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the pseudoenzyme PDX1.2 in complex with its cognate enzyme PDX1.3: a total eclipse.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
2ZJK
 
 | |
7F57
 
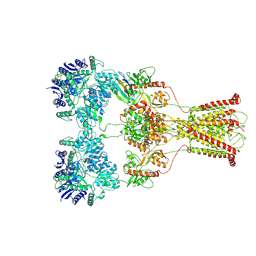 | | Kainate-bound GluK2-1xNeto2 complex, at the desensitized state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, kainate 2, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
7LVQ
 
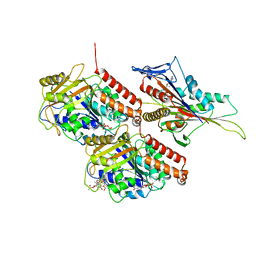 | | KIF14[391-743] - AMP-PNP closed state class in complex with a microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein KIF14, ... | | Authors: | Benoit, M.P.M.H, Asenjo, A.B, Paydar, M, Dhakal, S, Kwok, B, Sosa, H. | | Deposit date: | 2021-02-26 | | Release date: | 2021-05-05 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of mechano-chemical coupling by the mitotic kinesin KIF14.
Nat Commun, 12, 2021
|
|
7F59
 
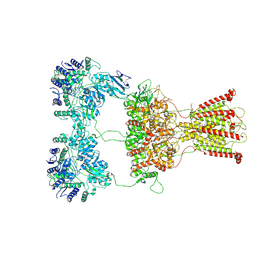 | | DNQX-bound GluK2-1xNeto2 complex | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | He, L.L, Gao, Y.W, Li, B, Zhao, Y. | | Deposit date: | 2021-06-21 | | Release date: | 2021-09-29 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Kainate receptor modulation by NETO2.
Nature, 599, 2021
|
|
