8DXJ
 
 | | HIV-1 reverse transcriptase/rilpivirine with bound fragment 1-N-methyl-4-(trifluoromethyl)benzene-1,2-diamine at the NNRTI adjacent site | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Chopra, A, Ruiz, F.X, Bauman, J.D, Arnold, E. | | Deposit date: | 2022-08-02 | | Release date: | 2023-05-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Halo Library, a Tool for Rapid Identification of Ligand Binding Sites on Proteins Using Crystallographic Fragment Screening.
J.Med.Chem., 66, 2023
|
|
1IHN
 
 | |
3DXJ
 
 | | Crystal structure of thermus thermophilus rna polymerase holoenzyme in complex with the antibiotic myxopyronin | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BACTERIAL RNA POLYMERASE BETA SUBUNIT; CHAIN C, M, ... | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2008-07-24 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The RNA polymerase "switch region" is a target for inhibitors.
Cell(Cambridge,Mass.), 135, 2008
|
|
3IYD
 
 | | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Catabolite gene activator, DNA (98-MER), ... | | Authors: | Hudson, B.P, Quispe, J, Lara, S, Kim, Y, Berman, H, Arnold, E, Ebright, R.H, Lawson, C.L. | | Deposit date: | 2009-08-01 | | Release date: | 2009-11-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (19.799999 Å) | | Cite: | Three-dimensional EM structure of an intact activator-dependent transcription initiation complex
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3DLK
 
 | | Crystal Structure of an engineered form of the HIV-1 Reverse Transcriptase, RT69A | | Descriptor: | Reverse transcriptase/ribonuclease H, SULFATE ION, p51 RT | | Authors: | Ho, W.C, Bauman, J.D, Himmel, D.M, Das, K, Arnold, E. | | Deposit date: | 2008-06-27 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal engineering of HIV-1 reverse transcriptase for structure-based drug design.
Nucleic Acids Res., 36, 2008
|
|
3S24
 
 | | Crystal structure of human mRNA guanylyltransferase | | Descriptor: | SULFATE ION, mRNA-capping enzyme | | Authors: | Das, K, Chu, C, Thyminski, J.R, Bauman, J.D, Guan, R, Qiu, W, Montelione, G.T, Arnold, E, Shatkin, A.J. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.0137 Å) | | Cite: | Structure of the guanylyltransferase domain of human mRNA capping enzyme.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
7KWU
 
 | | Crystal Structure of HIV-1 RT in Complex with 16c (K07-15) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-{[4-(4-cyano-2,6-dimethylphenoxy)-5-(pyridin-4-yl)pyrimidin-2-yl]amino}piperidin-1-yl)methyl]benzamide, MAGNESIUM ION, ... | | Authors: | Ruiz, F.X, Arnold, E. | | Deposit date: | 2020-12-02 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | 2,4,5-Trisubstituted Pyrimidines as Potent HIV-1 NNRTIs: Rational Design, Synthesis, Activity Evaluation, and Crystallographic Studies.
J.Med.Chem., 64, 2021
|
|
6VRP
 
 | |
6VUP
 
 | |
6VUN
 
 | | Reverse Transcriptase Diabody with R83C Mutation | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Single-chain Fv | | Authors: | Chesterman, C, Arnold, E. | | Deposit date: | 2020-02-16 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Co-crystallization with diabodies: A case study for the introduction of synthetic symmetry.
Structure, 29, 2021
|
|
6VUO
 
 | |
6VUG
 
 | |
2NCK
 
 | |
3KLF
 
 | | Crystal structure of wild-type HIV-1 Reverse Transcriptase crosslinked to a DSDNA with a bound excision product, AZTPPPPA | | Descriptor: | DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(2DA))-3'), DNA (5'-D(*AP*T*GP*CP*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), GLYCEROL, ... | | Authors: | Tu, X, Das, K, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KLE
 
 | | Crystal structure of AZT-resistant HIV-1 Reverse Transcriptase crosslinked to a DSDNA with a bound excision product, AZTPPPPA | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(2DA))-3'), GLYCEROL, ... | | Authors: | Tu, X, Das, K, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3K2P
 
 | | HIV-1 Reverse Transcriptase Isolated RnaseH Domain with the Inhibitor beta-thujaplicinol Bound at the Active Site | | Descriptor: | 2,7-dihydroxy-4-(propan-2-yl)cyclohepta-2,4,6-trien-1-one, MANGANESE (II) ION, Reverse Transcriptase | | Authors: | Pauly, T.A, Himmel, D.M, Maegley, K, Arnold, E. | | Deposit date: | 2009-09-30 | | Release date: | 2010-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of HIV-1 reverse transcriptase with the inhibitor beta-Thujaplicinol bound at the RNase H active site.
Structure, 17, 2009
|
|
5D3G
 
 | | Structure of HIV-1 Reverse Transcriptase Bound to a Novel 38-mer Hairpin Template-Primer DNA Aptamer | | Descriptor: | DNA aptamer (38-MER), GLYCEROL, HIV-1 REVERSE TRANSCRIPTASE P51 subunit, ... | | Authors: | Miller, M.T, Tuske, S, Das, K, Arnold, E. | | Deposit date: | 2015-08-06 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of HIV-1 reverse transcriptase bound to a novel 38-mer hairpin template-primer DNA aptamer.
Protein Sci., 25, 2016
|
|
5CYM
 
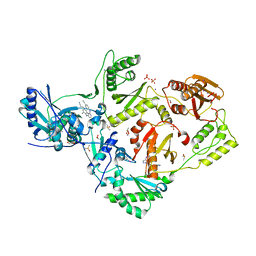 | | HIV-1 reverse transcriptase complexed with 4-iodopyrazole | | Descriptor: | 1,2-ETHANEDIOL, 4-IODOPYRAZOLE, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Bauman, J.D, Arnold, E. | | Deposit date: | 2015-07-30 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Rapid experimental SAD phasing and hot spot identification with halogenated fragments
Iucrj, 3, 2016
|
|
5CXR
 
 | |
5CYQ
 
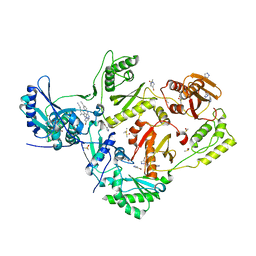 | | HIV-1 reverse transcriptase complexed with 4-bromopyrazole | | Descriptor: | 4-bromo-1H-pyrazole, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, BROMIDE ION, ... | | Authors: | Bauman, J.D, Arnold, E. | | Deposit date: | 2015-07-30 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.147 Å) | | Cite: | Rapid experimental SAD phasing and hot-spot identification with halogenated fragments.
IUCrJ, 3, 2016
|
|
5CW1
 
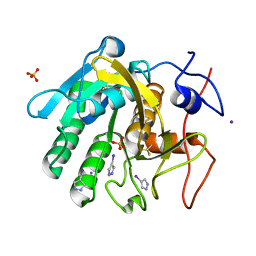 | | Proteinase K complexed with 4-iodopyrazole | | Descriptor: | 4-IODOPYRAZOLE, IODIDE ION, Proteinase K, ... | | Authors: | Bauman, J.D, Arnold, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-12-30 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Rapid experimental SAD phasing and hot spot identification with halogenated fragments
Iucrj, 3, 2016
|
|
3KLI
 
 | |
3KLG
 
 | | Crystal structure of AZT-resistant HIV-1 Reverse Transcriptase crosslinked to pre-translocation AZTMP-Terminated DNA (COMPLEX N) | | Descriptor: | DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*AP*(ATM))-3'), DNA (5'-D(*AP*T*GP*CP*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), Reverse transcriptase/ribonuclease H, ... | | Authors: | Tu, X, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3KLH
 
 | | Crystal structure of AZT-Resistant HIV-1 Reverse Transcriptase crosslinked to post-translocation AZTMP-Terminated DNA (COMPLEX P) | | Descriptor: | DNA (5'-D(*AP*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*T*GP*CP*TP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), MAGNESIUM ION, ... | | Authors: | Tu, X, Sarafianos, S.G, Arnold, E. | | Deposit date: | 2009-11-07 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of HIV-1 resistance to AZT by excision.
Nat.Struct.Mol.Biol., 17, 2010
|
|
1KXG
 
 | | The 2.0 Ang Resolution Structure of BLyS, B Lymphocyte Stimulator. | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, B lymphocyte stimulator, CITRIC ACID, ... | | Authors: | Oren, D.A, Li, Y, Volovik, Y, Morris, T.S, Dharia, C, Das, K, Galperina, O, Gentz, R, Arnold, E. | | Deposit date: | 2002-01-31 | | Release date: | 2002-03-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of BLyS receptor recognition.
Nat.Struct.Biol., 9, 2002
|
|
