1YB3
 
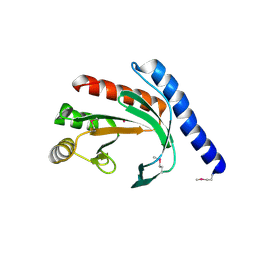 | | Conserved hypothetical protein Pfu-178653-001 from Pyrococcus furiosus | | Descriptor: | UNKNOWN ATOM OR ION, hypothetical protein | | Authors: | Habel, J, Zhou, W, Chang, J, Zhao, M, Xu, H, Chen, L, Lee, D, Nguyen, D, Chang, S.-H, Horanyi, P, Florence, Q, Tempel, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-19 | | Release date: | 2005-02-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Conserved hypothetical protein Pfu-178653-001 from Pyrococcus furiosus
To be published
|
|
1YBX
 
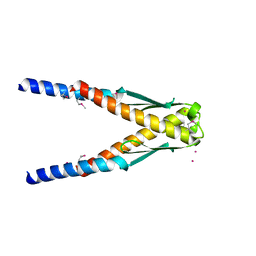 | | Conserved hypothetical protein Cth-383 from Clostridium thermocellum | | Descriptor: | Conserved hypothetical protein, UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Chang, J, Zhao, M, Habel, J, Kataeva, I, Xu, H, Chen, L, Lee, D, Nguyen, J, Chang, S.-H, Horanyi, P, Florence, Q, Zhou, W, Lin, D, Zhang, H, Ljundahl, L, Liu, Z.-J, Rose, J, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Conserved hypothetical protein Cth-383 from Clostridium thermocellum
To be published
|
|
1YD7
 
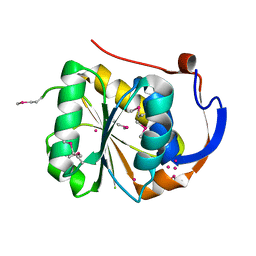 | | Conserved hypothetical protein Pfu-1647980-001 from Pyrococcus furiosus | | Descriptor: | 2-keto acid:ferredoxin oxidoreductase subunit alpha, UNKNOWN ATOM OR ION | | Authors: | Horanyi, P, Florence, Q, Zhou, W, Chang, J, Zhao, M, Habel, J, Xu, H, Chen, L, Lee, D, Nguyen, D, Chang, S.-H, Tempel, W, Lin, D, Zhang, H, Praissman, J, Jenney Jr, F.E, Adams, M.W.W, Liu, Z.-J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-23 | | Release date: | 2005-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conserved hypothetical protein Pfu-1647980-001 from Pyrococcus furiosus
To be published
|
|
4BH6
 
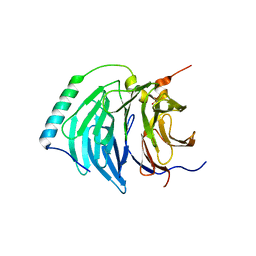 | | Insights into degron recognition by APC coactivators from the structure of an Acm1-Cdh1 complex | | Descriptor: | APC/C ACTIVATOR PROTEIN CDH1, APC/C-CDH1 MODULATOR 1 | | Authors: | He, J, Chao, W.C.H, Zhang, Z, Yang, J, Cronin, N, Barford, D. | | Deposit date: | 2013-03-29 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights Into Degron Recognition by Apc/C Coactivators from the Structure of an Acm1-Cdh1 Complex.
Mol.Cell, 50, 2013
|
|
8ISK
 
 | | Pr conformer of Zea mays phytochrome A1 - ZmphyA1-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
8ISJ
 
 | | Pr conformer of Arabidopsis thaliana phytochrome A - AtphyA-Pr | | Descriptor: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | Authors: | Zhang, Y, Ma, C, Zhao, J, Gao, N, Wang, J. | | Deposit date: | 2023-03-20 | | Release date: | 2023-08-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into plant phytochrome A as a highly sensitized photoreceptor.
Cell Res., 33, 2023
|
|
1XI8
 
 | | Molybdenum cofactor biosynthesis protein from Pyrococcus furiosus Pfu-1657500-001 | | Descriptor: | Molybdenum cofactor biosynthesis protein | | Authors: | Zhou, W, Zhao, M, Chang, J.C, Liu, Z.-J, Chen, L, Horanyi, P, Xu, H, Yang, H, Habel, J.E, Lee, D, Chang, S.-H, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Molybdenum cofactor biosynthesis protein from Pyrococcus furiosus Pfu-1657500-001
To be published
|
|
7VID
 
 | |
7XZR
 
 | | Crystal structure of TNIK-AMPPNP-thiopeptide TP15 complex | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SULFATE ION, ... | | Authors: | Hamada, K, Vinogradov, A.A, Zhang, Y, Chang, J.S, Nishimura, H, Goto, Y, Onaka, H, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | De Novo Discovery of Thiopeptide Pseudo-natural Products Acting as Potent and Selective TNIK Kinase Inhibitors.
J.Am.Chem.Soc., 144, 2022
|
|
7XZQ
 
 | | Crystal structure of TNIK-thiopeptide TP1 complex | | Descriptor: | 1,4-BUTANEDIOL, TRAF2 and NCK-interacting protein kinase, thiopeptide TP1 | | Authors: | Hamada, K, Vinogradov, A.A, Zhang, Y, Chang, J.S, Nishimura, H, Goto, Y, Onaka, H, Suga, H, Ogata, K, Sengoku, T. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | De Novo Discovery of Thiopeptide Pseudo-natural Products Acting as Potent and Selective TNIK Kinase Inhibitors.
J.Am.Chem.Soc., 144, 2022
|
|
7D1D
 
 | | Crystal structure of Bacteroides thetaiotaomicron glutaminyl cyclase bound to 1-benzylimidazole | | Descriptor: | 1-BENZYL-1H-IMIDAZOLE, Glutamine cyclotransferase, ZINC ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D2B
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with a Ni ion bound to the active site | | Descriptor: | Glutaminyl-peptide cyclotransferase, NICKEL (II) ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D18
 
 | | Crystal structure of Acidobacteriales bacterium glutaminyl cyclase | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Peptidase M28, ... | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D2I
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with a Fe ion bound to the active site | | Descriptor: | FE (III) ION, Glutaminyl-peptide cyclotransferase, SULFATE ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D23
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with one K ion bound to the active site | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glutaminyl-peptide cyclotransferase, POTASSIUM ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-15 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D2D
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with a Mn ion bound to the active site | | Descriptor: | Glutaminyl-peptide cyclotransferase, MANGANESE (II) ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-16 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7DKG
 
 | | Influenza H5N1 nucleoprotein (truncated) in complex with nucleotides | | Descriptor: | Nucleoprotein, RNA (5'-R(P*(OMU)P*(OMU)P*(OMU))-3') | | Authors: | Tang, Y.S, Xu, S, Chen, Y.W, Wang, J.H, Shaw, P.C. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of influenza nucleoprotein complexed with nucleic acid provide insights into the mechanism of RNA interaction.
Nucleic Acids Res., 49, 2021
|
|
7D17
 
 | | Crystal structure of Macrostomum lignano glutaminyl cyclase | | Descriptor: | Glutaminyl-peptide cyclotransferase, ZINC ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D1Y
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with two Co ions bound to the active site | | Descriptor: | COBALT (II) ION, Glutaminyl-peptide cyclotransferase | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-15 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D1P
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with three Cd ions bound to the active site | | Descriptor: | BICARBONATE ION, CADMIUM ION, Glutaminyl-peptide cyclotransferase | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-15 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D21
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with two Zn ions bound to the active site | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Glutaminyl-peptide cyclotransferase, ZINC ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-15 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D1N
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with three Cu ions bound to the active site | | Descriptor: | BICARBONATE ION, COPPER (II) ION, Glutaminyl-peptide cyclotransferase | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-15 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D1E
 
 | | Crystal structure of Bacteroides thetaiotaomicron glutaminyl cyclase bound to N-acetylhistamine | | Descriptor: | Leucine aminopeptidase, N-[2-(1H-IMIDAZOL-4-YL)ETHYL]ACETAMIDE, ZINC ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D1H
 
 | | Crystal structure of Ixodes scapularis glutaminyl cyclase with D238A mutation | | Descriptor: | Glutaminyl-peptide cyclotransferase, ZINC ION | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
7D1B
 
 | | Crystal structure of Fimbriiglobus ruber glutaminyl cyclase | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Huang, K.-F, Huang, J.-S, Wu, M.-L, Hsieh, W.-L, Wang, A.H.-J. | | Deposit date: | 2020-09-14 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | A Unique Carboxylic-Acid Hydrogen-Bond Network (CAHBN) Confers Glutaminyl Cyclase Activity on M28 Family Enzymes.
J.Mol.Biol., 433, 2021
|
|
