1KUX
 
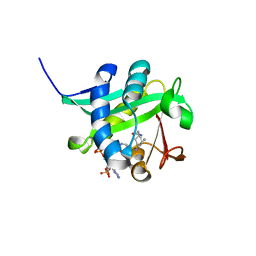 | | X-ray Crystallographic Studies of Serotonin N-acetyltransferase Catalysis and Inhibition | | Descriptor: | COA-S-TRIMETHYLENE-ACETYL-TRYPTAMINE, Serotonin N-acetyltransferase | | Authors: | Wolf, E, De Angelis, J, Khalil, E.M, Cole, P.A, Burley, S.K. | | Deposit date: | 2002-01-22 | | Release date: | 2002-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic studies of serotonin N-acetyltransferase catalysis and inhibition.
J.Mol.Biol., 317, 2002
|
|
3V7E
 
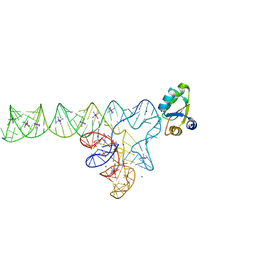 | | Crystal structure of YbxF bound to the SAM-I riboswitch aptamer | | Descriptor: | COBALT HEXAMMINE(III), MAGNESIUM ION, Ribosome-associated protein L7Ae-like, ... | | Authors: | Baird, N.J, Zhang, J, Hamma, T, Ferre-D'Amare, A.R. | | Deposit date: | 2011-12-21 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | YbxF and YlxQ are bacterial homologs of L7Ae and bind K-turns but not K-loops.
Rna, 18, 2012
|
|
4GO1
 
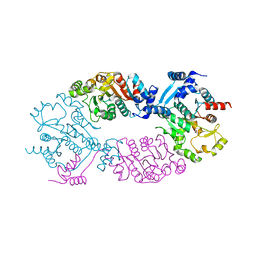 | | Crystal Structure of full length transcription repressor LsrR from E. coli. | | Descriptor: | GLYCEROL, Transcriptional regulator LsrR | | Authors: | Wu, M, Tao, Y, Liu, X, Zang, J. | | Deposit date: | 2012-08-17 | | Release date: | 2013-04-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Phosphorylated Autoinducer-2 Modulation of the Oligomerization State of the Global Transcription Regulator LsrR from Escherichia coli
J.Biol.Chem., 288, 2013
|
|
7VDR
 
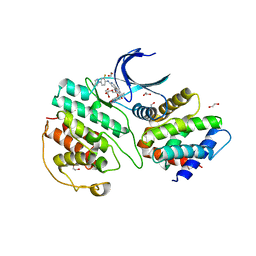 | | The structure of cyclin-dependent kinase 5 (CDK5) in complex with p25 and Compound 13 | | Descriptor: | (1R)-1-[7-[(2-fluoranyl-4-pyrazol-1-yl-phenyl)amino]-1,6-naphthyridin-2-yl]-1-(1-methylpiperidin-4-yl)ethanol, 1,2-ETHANEDIOL, Cyclin-dependent kinase 5 activator 1, ... | | Authors: | Malojcic, G, Clugston, S.L, Daniels, M, Harmange, J.C, Ledeborer, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and Optimization of Highly Selective Inhibitors of CDK5.
J.Med.Chem., 65, 2022
|
|
7VDP
 
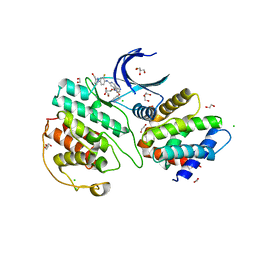 | | The structure of cyclin-dependent kinase 5 (CDK5) in complex with p25 and Compound 1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cyclin-dependent kinase 5 activator 1, ... | | Authors: | Malojcic, G, Clugston, S.L, Daniels, M, Harmange, J.C, Ledeborer, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery and Optimization of Highly Selective Inhibitors of CDK5.
J.Med.Chem., 65, 2022
|
|
7VDU
 
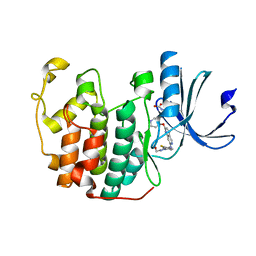 | | The structure of cyclin-dependent kinase 2 (CDK2) in complex with Compound 1 | | Descriptor: | Cyclin-dependent kinase 2, [1-[3-fluoranyl-4-[(2-piperidin-4-yloxy-1,6-naphthyridin-7-yl)amino]phenyl]pyrazol-3-yl]methanol | | Authors: | Malojcic, G, Clugston, S.L, Daniels, M, Harmange, J.C, Ledeborer, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovery and Optimization of Highly Selective Inhibitors of CDK5.
J.Med.Chem., 65, 2022
|
|
6TM1
 
 | |
1NPE
 
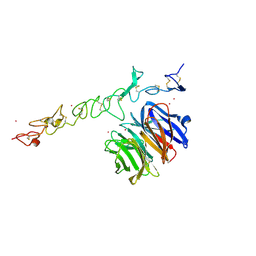 | | Crystal structure of Nidogen/Laminin Complex | | Descriptor: | CADMIUM ION, Laminin gamma-1 chain, nidogen | | Authors: | Takagi, J, Yang, Y.T, Liu, J.-H, Wang, J.-H, Springer, T.A. | | Deposit date: | 2003-01-17 | | Release date: | 2003-08-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Complex between nidogen and laminin fragments reveals a paradigmatic
beta-propeller interface
Nature, 424, 2003
|
|
3EXF
 
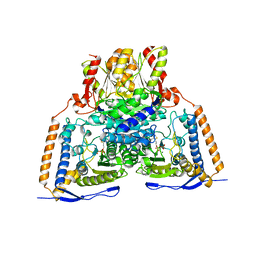 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
3J1Z
 
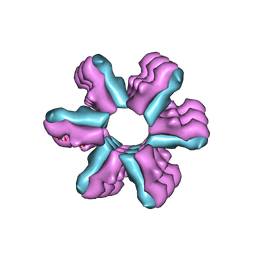 | | Inward-Facing Conformation of the Zinc Transporter YiiP revealed by Cryo-electron Microscopy | | Descriptor: | Cation efflux family protein | | Authors: | Coudray, N, Valvo, S, Hu, M, Lasala, R, Kim, C, Vink, M, Zhou, M, Provasi, D, Filizola, M, Tao, J, Fang, J, Penczek, P.A, Ubarretxena-Belandia, I, Stokes, D.L, Transcontinental EM Initiative for Membrane Protein Structure (TEMIMPS) | | Deposit date: | 2012-07-24 | | Release date: | 2012-10-10 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (13 Å) | | Cite: | Inward-facing conformation of the zinc transporter YiiP revealed by cryoelectron microscopy.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3EXG
 
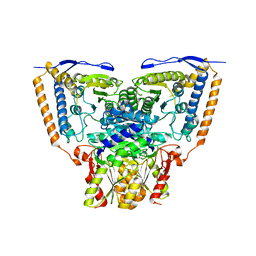 | | Crystal structure of the pyruvate dehydrogenase (E1p) component of human pyruvate dehydrogenase complex | | Descriptor: | POTASSIUM ION, Pyruvate dehydrogenase E1 component subunit alpha, somatic form, ... | | Authors: | Kato, M, Wynn, R.M, Chuang, J.L, Tso, S.-C, Machius, M, Li, J, Chuang, D.T. | | Deposit date: | 2008-10-16 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.011 Å) | | Cite: | Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Structure, 16, 2008
|
|
7XV3
 
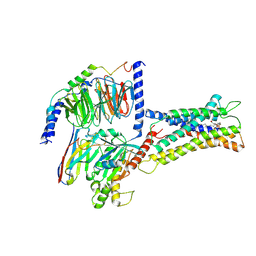 | | Cryo-EM structure of LPS-bound GPR174 in complex with Gs protein | | Descriptor: | (2~{S})-2-$l^{4}-azanyl-3-[[(2~{R})-3-octadecanoyloxy-2-oxidanyl-propoxy]-oxidanyl-oxidanylidene-$l^{6}-phosphanyl]oxy-propanoic acid, Engineered G protein subunit S (mini-Gs), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Liang, J. | | Deposit date: | 2022-05-20 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis of lysophosphatidylserine receptor GPR174 ligand recognition and activation.
Nat Commun, 14, 2023
|
|
2P5S
 
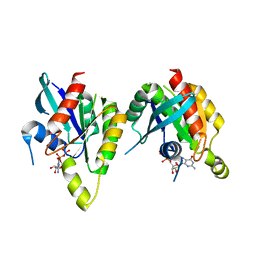 | | RAB domain of human RASEF in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, RAS and EF-hand domain containing, UNKNOWN ATOM OR ION | | Authors: | Zhu, H, Wang, J, Shen, Y, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-03-16 | | Release date: | 2007-03-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | RAB domain of human RASEF in complex with GDP
To be Published
|
|
3JB3
 
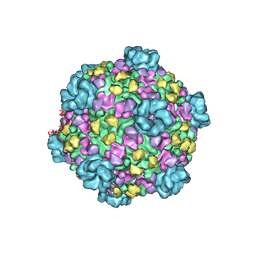 | | Atomic model of cytoplasmic polyhedrosis virus with SAM, GTP and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Capsid protein VP1, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Yu, X.K, Jiang, J.S, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2015-07-06 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A putative ATPase mediates RNA transcription and capping in a dsRNA virus.
Elife, 4, 2015
|
|
3QEP
 
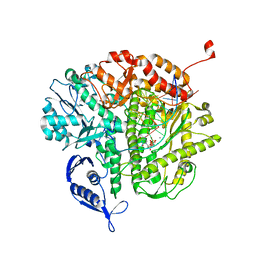 | | RB69 DNA Polymerase (L561A/S565G/Y567A) Ternary Complex with dTTP Opposite Difluorotoluene Nucleoside | | Descriptor: | CALCIUM ION, DNA (5'-D(*GP*CP*GP*GP*AP*CP*TP*GP*CP*TP*TP*AP*(DOC))-3'), DNA (5'-D(*TP*CP*GP*(DFT)P*GP*TP*AP*AP*GP*CP*AP*GP*TP*CP*CP*GP*CP*G)-3'), ... | | Authors: | Xia, S, Konigsberg, W.H, Wang, J. | | Deposit date: | 2011-01-20 | | Release date: | 2011-08-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hydrogen-bonding capability of a templating difluorotoluene nucleotide residue in an RB69 DNA polymerase ternary complex.
J.Am.Chem.Soc., 133, 2011
|
|
7WKC
 
 | |
6IJZ
 
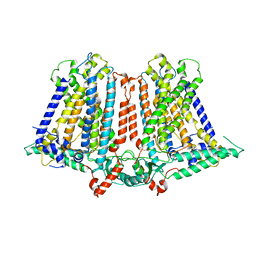 | | Structure of a plant cation channel | | Descriptor: | Calcium permeable stress-gated cation channel 1 | | Authors: | Sun, L, Wang, J, Liu, X. | | Deposit date: | 2018-10-12 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structure of the hyperosmolality-gated calcium-permeable channel OSCA1.2.
Nat Commun, 9, 2018
|
|
5XVD
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an air-oxidized condition | | Descriptor: | FE3-S4 CLUSTER, FE4-S4-O CLUSTER, GLYCEROL, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
3K6U
 
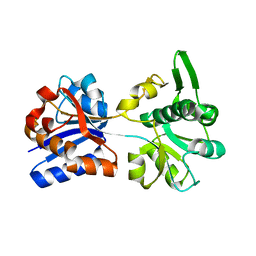 | | M. acetivorans Molybdate-Binding Protein (ModA) in Unliganded Open Form | | Descriptor: | Solute-binding protein MA_0280 | | Authors: | Chan, S, Giuroiu, I, Chernishof, I, Sawaya, M.R, Chiang, J, Gunsalus, R.P, Arbing, M.A, Perry, L.J. | | Deposit date: | 2009-10-09 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Apo and ligand-bound structures of ModA from the archaeon Methanosarcina acetivorans
Acta Crystallogr.,Sect.F, 66, 2010
|
|
7EO7
 
 | | Crystal structure of HCoV-NL63 3C-like protease in complex with an inhibitor Shikonin | | Descriptor: | 2-[(1R)-4-methyl-1-oxidanyl-pent-3-enyl]-5,8-bis(oxidanyl)naphthalene-1,4-dione, 3C-like proteinase | | Authors: | Gao, H.X, Zhang, Y.T, Zhou, X.L, Zhong, F.L, Li, J, Zhang, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.24916625 Å) | | Cite: | Structure-Based Discovery and Structural Basis of a Novel Broad-Spectrum Natural Product against the Main Protease of Coronavirus.
J.Virol., 96, 2022
|
|
7KGB
 
 | |
7LX0
 
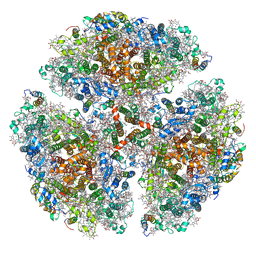 | | Quantitative assessment of chlorophyll types in cryo-EM maps of photosystem I acclimated to far-red light | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Gisriel, C.J, Wang, J. | | Deposit date: | 2021-03-02 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Quantitative assessment of chlorophyll types in cryo-EM maps of photosystem I acclimated to far-red light
BBA Adv, 1, 2021
|
|
2GP5
 
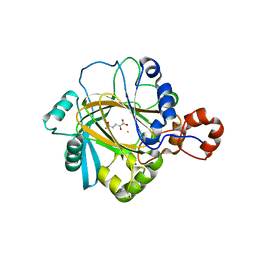 | | Crystal structure of catalytic core domain of jmjd2A complexed with alpha-Ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, Jumonji domain-containing protein 2A, ... | | Authors: | Chen, Z, Zang, J, Whetstine, J, Hong, X, Davrazou, F, Kutateladze, T.G, Simpson, M, Dai, S, Hagman, J, Shi, Y, Zhang, G. | | Deposit date: | 2006-04-16 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural insights into histone demethylation by JMJD2 family members
Cell(Cambridge,Mass.), 125, 2006
|
|
6GH2
 
 | | Paenibacillus sp. YM1 laminaribiose phosphorylase with alpha-glc-1-phosphate bound | | Descriptor: | 1,2-ETHANEDIOL, 1-O-phosphono-alpha-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Kuhaudomlarp, S, Walpole, S, Stevenson, C.E.M, Nepogodiev, S.A, Lawson, D.M, Angulo, J, Field, R.A. | | Deposit date: | 2018-05-04 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Unravelling the Specificity of Laminaribiose Phosphorylase from Paenibacillus sp. YM-1 towards Donor Substrates Glucose/Mannose 1-Phosphate by Using X-ray Crystallography and Saturation Transfer Difference NMR Spectroscopy.
Chembiochem, 20, 2019
|
|
2GP3
 
 | | Crystal structure of the catalytic core domain of jmjd2a | | Descriptor: | FE (II) ION, Jumonji domain-containing protein 2A, ZINC ION | | Authors: | Chen, Z, Zang, J, Whetstine, J, Hong, X, Davrazou, F, Kutateladze, T.G, Simpson, M, Dai, S, Hagman, J, Shi, Y, Zhang, G. | | Deposit date: | 2006-04-16 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into histone demethylation by JMJD2 family members
Cell(Cambridge,Mass.), 125, 2006
|
|
