5XZ7
 
 | | Crystal Structure of Phosphofructokinase from Staphylococcus aureus in complex with adenylylimidodiphosphate, the ATP analogue | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ATP-dependent 6-phosphofructokinase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wang, C.L, Tian, T, Zang, J.Y. | | Deposit date: | 2017-07-11 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Insights into the Regulation of Staphylococcus aureus Phosphofructokinase by Tetramer-Dimer Conversion.
Biochemistry, 57, 2018
|
|
5Y0V
 
 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with berberine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BERBERINE, ... | | Authors: | Duan, Y.W, Liu, T, Tang, J.Y, Li, M, Yang, Q. | | Deposit date: | 2017-07-18 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | Glycoside hydrolase family 18 and 20 enzymes are novel targets of the traditional medicine berberine.
J. Biol. Chem., 293, 2018
|
|
6WQZ
 
 | | Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery | | Descriptor: | Autophagy-related protein 9A, Lauryl Maltose Neopentyl Glycol | | Authors: | Guardia, C.M, Tan, X, Lian, T, Rana, M.S, Zhou, W, Christenson, E.T, Lowry, A.J, Faraldo-Gomez, J.D, Bonifacino, J.S, Jiang, J, Banerjee, A. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of Human ATG9A, the Only Transmembrane Protein of the Core Autophagy Machinery.
Cell Rep, 31, 2020
|
|
7Y78
 
 | | Crystal structure of Cry78Aa | | Descriptor: | 1,2-ETHANEDIOL, AMMONIUM ION, Toxin | | Authors: | Cao, B.B, Nie, Y.F, Wang, N.C, Guan, Z.Y, Zhang, D.L, Zhang, J. | | Deposit date: | 2022-06-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of Cry78Aa from Bacillus thuringiensis provides insights into its insecticidal activity.
Commun Biol, 5, 2022
|
|
7XU2
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU0
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-211 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XTZ
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-1 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU1
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-122 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU4
 
 | | Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU5
 
 | | Structure of SARS-CoV-2 D614G Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU3
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7XU6
 
 | | Structure of SARS-CoV-2 Spike Protein with Engineered x3 Disulfide (x3(D427C, V987C) and single Arg S1/S2 cleavage site), incubated in Low pH after 40-Day Storage in PBS, Locked-2 Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, ... | | Authors: | Qu, K, Chen, Q, Ciazynska, K.A, Liu, B, Zhang, X, Wang, J, He, Y, Guan, J, He, J, Liu, T, Zhang, X, Carter, A.P, Xiong, X, Briggs, J.A.G. | | Deposit date: | 2022-05-18 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Engineered disulfide reveals structural dynamics of locked SARS-CoV-2 spike.
Plos Pathog., 18, 2022
|
|
7DAC
 
 | | Human RIPK3 amyloid fibril revealed by solid-state NMR | | Descriptor: | Receptor-interacting serine/threonine-protein kinase 3 | | Authors: | Wu, X.L, Zhang, J, Dong, X.Q, Liu, J, Li, B, Hu, H, Wang, J, Wang, H.Y, Lu, J.X. | | Deposit date: | 2020-10-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | The structure of a minimum amyloid fibril core formed by necroptosis-mediating RHIM of human RIPK3.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7D72
 
 | | Cryo-EM structures of human GMPPA/GMPPB complex bound to GDP-Mannose | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, MAGNESIUM ION, Mannose-1-phosphate guanyltransferase alpha, ... | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, Cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2021-06-02 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7D73
 
 | | Cryo-EM structure of GMPPA/GMPPB complex bound to GTP (State I) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, GUANOSINE-5'-TRIPHOSPHATE, Mannose-1-phosphate guanyltransferase alpha, ... | | Authors: | Zheng, L, Liu, Z, Wang, Y, Yang, F, Wang, J, Qing, J, Cai, X, Mo, X, Gao, N, Jia, D. | | Deposit date: | 2020-10-02 | | Release date: | 2021-05-05 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-EM structures of human GMPPA-GMPPB complex reveal how cells maintain GDP-mannose homeostasis.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4DNV
 
 | | Crystal structure of the W285F mutant of UVB-resistance protein UVR8 | | Descriptor: | AT5g63860/MGI19_6 | | Authors: | Wu, D, Hu, Q, Yan, Z, Chen, W, Yan, C, Zhang, J, Wang, J, Shi, Y. | | Deposit date: | 2012-02-09 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Structural basis of ultraviolet-B perception by UVR8.
Nature, 484, 2012
|
|
8FE7
 
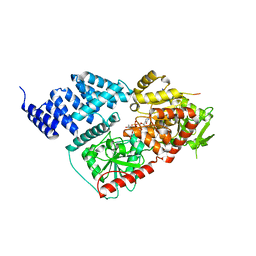 | |
8FE6
 
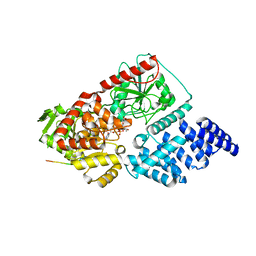 | |
8FUF
 
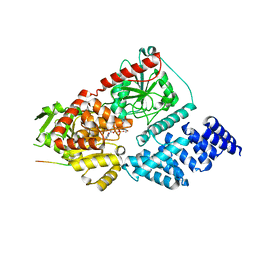 | |
5V9U
 
 | | Crystal Structure of small molecule ARS-1620 covalently bound to K-Ras G12C | | Descriptor: | (S)-1-{4-[6-chloro-8-fluoro-7-(2-fluoro-6-hydroxyphenyl)quinazolin-4-yl] piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | Janes, M.R, Zhang, J, Li, L.-S, Hansen, R, Peters, U, Guo, X, Chen, Y, Babbar, A, Firdaus, S.J, Feng, J, Chen, J.H, Li, S, Brehmer, D, Darjania, L, Li, S, Long, Y.O, Thach, C, Liu, Y, Zarieh, A, Ely, T, Kucharski, J.M, Kessler, L.V, Wu, T, Wang, Y, Yao, Y, Deng, X, Zarrinkar, P, Dashyant, D, Lorenzi, M.V, Hu-Lowe, D, Patricelli, M.P, Ren, P, Liu, Y. | | Deposit date: | 2017-03-23 | | Release date: | 2018-02-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Targeting KRAS Mutant Cancers with a Covalent G12C-Specific Inhibitor.
Cell, 172, 2018
|
|
5YBB
 
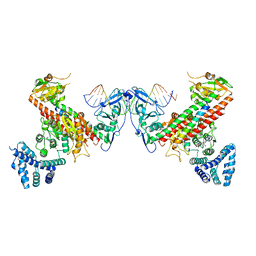 | | Structural basis underlying complex assembly andconformational transition of the type I R-M system | | Descriptor: | DNA, Restriction endonuclease S subunits, S-ADENOSYLMETHIONINE, ... | | Authors: | Liu, Y.P, Tang, Q, Zhang, J.Z, Tian, L.F, Gao, P, Yan, X.X. | | Deposit date: | 2017-09-04 | | Release date: | 2017-11-29 | | Last modified: | 2018-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis underlying complex assembly and conformational transition of the type I R-M system.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
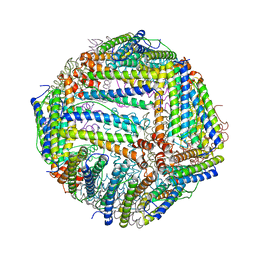
6J4A
 
 | |
3J9K
 
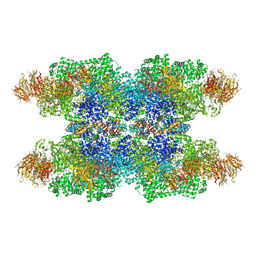 | | Structure of Dark apoptosome in complex with Dronc CARD domain | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Apaf-1 related killer DARK, Caspase Nc | | Authors: | Pang, Y, Bai, X, Yan, C, Hao, Q, Chen, Z, Wang, J, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-02-04 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the apoptosome: mechanistic insights into activation of an initiator caspase from Drosophila.
Genes Dev., 29, 2015
|
|
6LTP
 
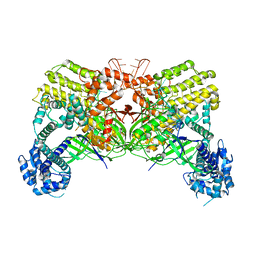 | | Crystal structure of Cas12i2 binary complex | | Descriptor: | Cas12i2, crRNA (56-mer RNA) | | Authors: | Huang, X, Sun, W, Cheng, Z, Chen, M, Li, X, Wang, J, Sheng, G, Gong, W, Wang, Y. | | Deposit date: | 2020-01-23 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Nat Commun, 11, 2020
|
|
6X4Y
 
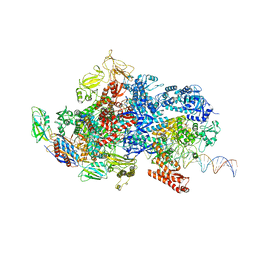 | | Mfd-bound E.coli RNA polymerase elongation complex - IV state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-24 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
