8K5I
 
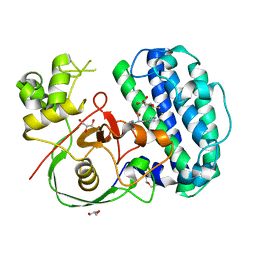 | | The structure of SenA in complex with N,N,N-trimethyl-histidine and thioglucose | | Descriptor: | 1-thio-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | Authors: | Liu, M, Yang, Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-21 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural insights into a novel nonheme iron-dependent oxygenase in selenoneine biosynthesis.
Int.J.Biol.Macromol., 256, 2023
|
|
8K5K
 
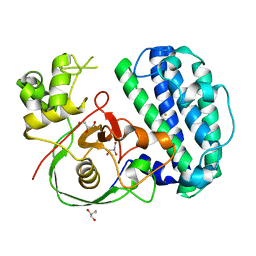 | | The structure of SenA | | Descriptor: | FE (III) ION, GLYCEROL, selenoneine synthase SenA | | Authors: | Liu, M, Yang, Y, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2023-07-21 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural insights into a novel nonheme iron-dependent oxygenase in selenoneine biosynthesis.
Int.J.Biol.Macromol., 256, 2023
|
|
7CAD
 
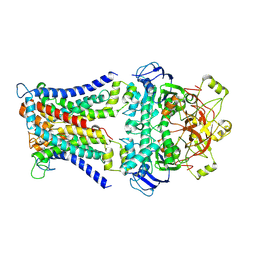 | | Mycobacterium smegmatis SugABC complex | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liu, F, Liang, J, Zhang, B, Gao, Y, Yang, X, Hu, T, Rao, Z. | | Deposit date: | 2020-06-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural basis of trehalose recycling by the ABC transporter LpqY-SugABC.
Sci Adv, 6, 2020
|
|
7YYH
 
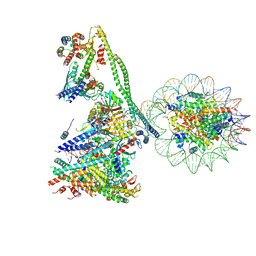 | | Structure of the human CCANdeltaT CENP-A alpha-satellite complex | | Descriptor: | Centromere protein C, Centromere protein H, Centromere protein I, ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Zhang, Z, Yang, J, Tischer, T, Predin, M, Dendooven, T, McLaughlin, S.H, Barford, D. | | Deposit date: | 2022-02-17 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
7YWX
 
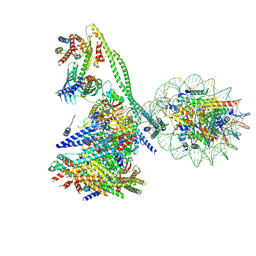 | | Structure of the human CCAN CENP-A alpha-satellite complex | | Descriptor: | Centromere protein C, Centromere protein H, Centromere protein I, ... | | Authors: | Yatskevich, S, Muir, K.W, Bellini, D, Zhang, Z, Yang, J, Tischer, T, Predin, M, Dendooven, T, McLaughlin, S.H, Barford, D. | | Deposit date: | 2022-02-14 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structure of the human inner kinetochore bound to a centromeric CENP-A nucleosome.
Science, 376, 2022
|
|
5HUG
 
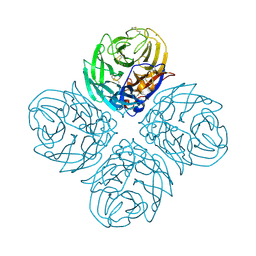 | | The crystal structure of neuraminidase from A/American green-winged teal/Washington/195750/2014 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yang, H, Carney, P.J, Guo, Z, Chang, J.C, Stevens, J. | | Deposit date: | 2016-01-27 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular Characterizations of Surface Proteins Hemagglutinin and Neuraminidase from Recent H5Nx Avian Influenza Viruses.
J.Virol., 90, 2016
|
|
7CAE
 
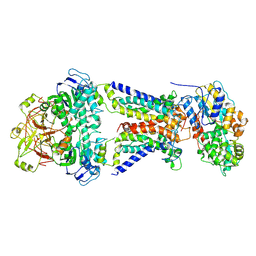 | | Mycobacterium smegmatis LpqY-SugABC complex in the resting state | | Descriptor: | ABC sugar transporter, permease component, ABC transporter, ... | | Authors: | Liu, F, Liang, J, Zhang, B, Gao, Y, Yang, X, Hu, T, Rao, Z. | | Deposit date: | 2020-06-08 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural basis of trehalose recycling by the ABC transporter LpqY-SugABC.
Sci Adv, 6, 2020
|
|
6L62
 
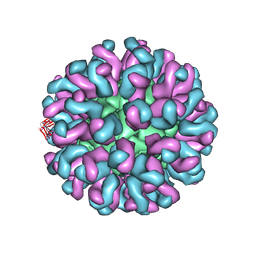 | | Neutralization mechanism of a monoclonal antibody targeting a porcine circovirus type 2 Cap protein conformational epitope | | Descriptor: | Capsid protein, Heavy chain of Fab fragment, Light chain of Fab fragment | | Authors: | Sun, Z, Huang, L, Xia, D, Wei, Y, Sun, E, Zhu, H, Bian, H, Wu, H, Feng, L, Wang, J, Liu, C. | | Deposit date: | 2019-10-25 | | Release date: | 2020-02-12 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Neutralization Mechanism of a Monoclonal Antibody Targeting a Porcine Circovirus Type 2 Cap Protein Conformational Epitope.
J.Virol., 94, 2020
|
|
5HUM
 
 | | The crystal structure of neuraminidase from A/Sichuan/26221/2014 influenza virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, neuraminidase | | Authors: | Yang, H, Carney, P.J, Guo, Z, Chang, J.C, Stevens, J. | | Deposit date: | 2016-01-27 | | Release date: | 2016-04-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular Characterizations of Surface Proteins Hemagglutinin and Neuraminidase from Recent H5Nx Avian Influenza Viruses.
J.Virol., 90, 2016
|
|
8EU3
 
 | | Cryo-EM structure of cGMP bound human CNGA3/CNGB3 channel in GDN, transition state 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel alpha-3, ... | | Authors: | Hu, Z, Zheng, X, Yang, J. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Conformational trajectory of allosteric gating of the human cone photoreceptor cyclic nucleotide-gated channel.
Nat Commun, 14, 2023
|
|
8ETP
 
 | | Cryo-EM structure of cGMP bound closed state of human CNGA3/CNGB3 channel in GDN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel alpha-3, ... | | Authors: | Hu, Z, Zheng, X, Yang, J. | | Deposit date: | 2022-10-17 | | Release date: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Conformational trajectory of allosteric gating of the human cone photoreceptor cyclic nucleotide-gated channel.
Nat Commun, 14, 2023
|
|
8EUC
 
 | | Cryo-EM structure of cGMP bound human CNGA3/CNGB3 channel in GDN, transition state 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYCLIC GUANOSINE MONOPHOSPHATE, Cyclic nucleotide-gated cation channel alpha-3, ... | | Authors: | Hu, Z, Zheng, X, Yang, J. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-09 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Conformational trajectory of allosteric gating of the human cone photoreceptor cyclic nucleotide-gated channel.
Nat Commun, 14, 2023
|
|
8EVA
 
 | |
8EVB
 
 | |
8EV9
 
 | |
8EVC
 
 | |
8EV8
 
 | |
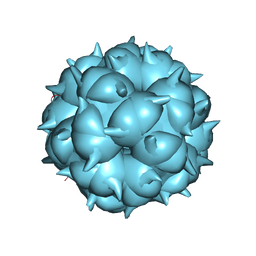
8TOC
 
 | | Acinetobacter phage AP205 | | Descriptor: | Coat protein, Maturation protein, RNA (4269-MER) | | Authors: | Meng, R, Xing, Z, Chang, J, Zhang, J. | | Deposit date: | 2023-08-03 | | Release date: | 2024-03-06 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Structural basis of Acinetobacter type IV pili targeting by an RNA virus.
Nat Commun, 15, 2024
|
|
6LU0
 
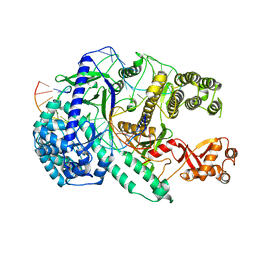 | | Crystal structure of Cas12i2 ternary complex with 12 nt spacer | | Descriptor: | Cas12i2, DNA (5'-D(*GP*CP*CP*GP*CP*TP*TP*TP*CP*TP*T)-3'), DNA (5'-D(*GP*CP*TP*TP*GP*CP*TP*CP*TP*GP*TP*TP*GP*AP*AP*AP*GP*CP*GP*GP*C)-3'), ... | | Authors: | Huang, X, Sun, W, Cheng, Z, Chen, M, Li, X, Wang, J, Sheng, G, Gong, W, Wang, Y. | | Deposit date: | 2020-01-24 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Nat Commun, 11, 2020
|
|
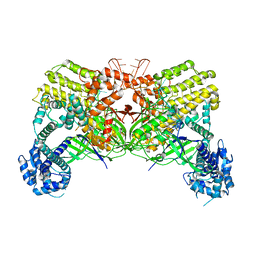
6LM3
 
 | | Neutralization mechanism of a monoclonal antibody targeting a porcine circovirus type 2 Cap protein conformational epitope | | Descriptor: | Capsid protein | | Authors: | Sun, Z, Huang, L, Xia, D, Wei, Y, Sun, E, Zhu, H, Bian, H, Wu, H, Feng, L, Wang, J, Liu, C. | | Deposit date: | 2019-12-24 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Neutralization Mechanism of a Monoclonal Antibody Targeting a Porcine Circovirus Type 2 Cap Protein Conformational Epitope.
J.Virol., 94, 2020
|
|
6LTP
 
 | | Crystal structure of Cas12i2 binary complex | | Descriptor: | Cas12i2, crRNA (56-mer RNA) | | Authors: | Huang, X, Sun, W, Cheng, Z, Chen, M, Li, X, Wang, J, Sheng, G, Gong, W, Wang, Y. | | Deposit date: | 2020-01-23 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for two metal-ion catalysis of DNA cleavage by Cas12i2.
Nat Commun, 11, 2020
|
|
6M5C
 
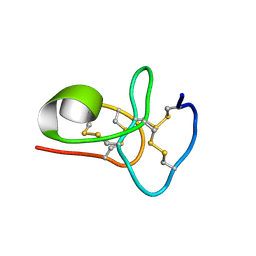 | | Solution structure of avenatide aV1 | | Descriptor: | avenatide aV1 | | Authors: | Tay, S.V, Wong, K.H, Huang, J.Y, Fan, J.S, Yang, D.W, Tam, J.P. | | Deposit date: | 2020-03-10 | | Release date: | 2021-03-10 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structure of avenatide aV1
To Be Published
|
|
8W8Q
 
 | | Cryo-EM structure of the GPR101-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Sun, J.P, Gao, N, Yu, X, Wang, G.P, Yang, F, Wang, J.Y, Yang, Z, Guan, Y. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
8W8S
 
 | | Cryo-EM structure of the AA14-bound GPR101 complex | | Descriptor: | 1-(4-methylpyridin-2-yl)-3-[3-(trifluoromethyl)phenyl]thiourea, Probable G-protein coupled receptor 101 | | Authors: | Sun, J.P, Yu, X, Gao, N, Yang, F, Wang, J.Y, Yang, Z, Guan, Y, Wang, G.P. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
8W8R
 
 | | Cryo-EM structure of the AA-14-bound GPR101-Gs complex | | Descriptor: | 1-(4-methylpyridin-2-yl)-3-[3-(trifluoromethyl)phenyl]thiourea, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Yu, X, Gao, N, Yang, F, Wang, J.Y, Yang, Z, Guan, Y, Wang, G.P. | | Deposit date: | 2023-09-04 | | Release date: | 2024-01-03 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of GPR101-Gs enables identification of ligands with rejuvenating potential.
Nat.Chem.Biol., 20, 2024
|
|
