6RV6
 
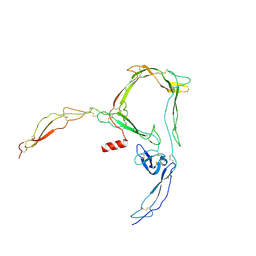 | | Structure of properdin lacking TSR3 based on anomalous data | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Properdin, alpha-D-mannopyranose, ... | | Authors: | Pedersen, D.V, Andersen, G.R. | | Deposit date: | 2019-05-31 | | Release date: | 2019-08-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.507 Å) | | Cite: | Structural Basis for Properdin Oligomerization and Convertase Stimulation in the Human Complement System.
Front Immunol, 10, 2019
|
|
2NPF
 
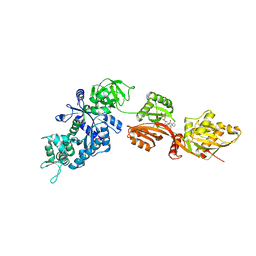 | | Structure of eEF2 in complex with moriniafungin | | Descriptor: | (1S,4R,5R,9S,11S)-2-({[(2S,5R,6R,7R,9S,10R)-2-(7-CARBOXYHEPTYL)-6-HYDROXY-10-METHOXY-9-METHYL-3-OXO-1,4,8-TRIOXASPIRO[4 .5]DEC-7-YL]OXY}METHYL)-9-FORMYL-13-ISOPROPYL-5-METHYLTETRACYCLO[7.4.0.02,11.04.8]TRIDEC-12-ENE-1-CARBOXYLIC ACID, Elongation factor 2, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Soe, R, Mosley, R.T, Andersen, G.R. | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Sordarin derivatives induce a novel conformation of the yeast ribosome translocation factor eEF2
J.Biol.Chem., 282, 2007
|
|
2E1R
 
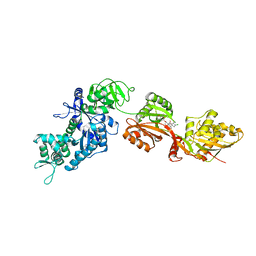 | | Structure of eEF2 in complex with a sordarin derivative | | Descriptor: | (1S,4S,5S,6R,9S,11S)-6-CHLORO-9-FORMYL-13-ISOPROPYL-5-METHYL-2-({[(3AR,5R,7R ,7AS)-7-METHYL-3-METHYLENEHEXAHYDRO-2H-FURO[2,3-C]PYRAN-5-YL]OXY}METHYL)TETR ACYCLO[7.4.0.02,11.04,8]TRIDEC-12-ENE-1-CARBOXYLIC ACI, Elongation factor 2, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Soe, R, Mosley, R.T, Andersen, G.R. | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Sordarin derivatives induce a novel conformation of the yeast ribosome translocation factor eEF2
J.Biol.Chem., 282, 2007
|
|
2P8Z
 
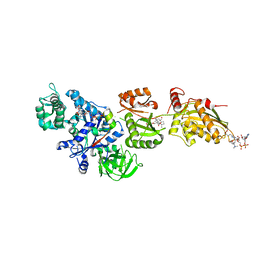 | | Fitted structure of ADPR-eEF2 in the 80S:ADPR-eEF2:GDPNP:sordarin cryo-EM reconstruction | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Elongation factor 2, Elongation factor Tu-B, ... | | Authors: | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
2P8W
 
 | | Fitted structure of eEF2 in the 80S:eEF2:GDPNP cryo-EM reconstruction | | Descriptor: | Elongation factor 2, Elongation factor Tu-B, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (11.3 Å) | | Cite: | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
2P8X
 
 | | Fitted structure of ADPR-eEF2 in the 80S:ADPR-eEF2:GDPNP cryo-EM reconstruction | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Elongation factor 2, Elongation factor Tu-B, ... | | Authors: | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (9.7 Å) | | Cite: | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
2P8Y
 
 | | Fitted structure of ADPR-eEF2 in the 80S:ADPR-eEF2:GDP:sordarin cryo-EM reconstruction | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Elongation factor 2, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Taylor, D.J, Nilsson, J, Merrill, A.R, Andersen, G.R, Nissen, P, Frank, J. | | Deposit date: | 2007-03-23 | | Release date: | 2007-05-08 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | Structures of modified eEF2.80S ribosome complexes reveal the role of GTP hydrolysis in translocation.
Embo J., 26, 2007
|
|
1N0U
 
 | | Crystal structure of yeast elongation factor 2 in complex with sordarin | | Descriptor: | Elongation factor 2, [1R-(1.ALPHA.,3A.BETA.,4.BETA.,4A.BETA.,7.BETA.,7A.ALPHA.,8A.BETA.)]8A-[(6-DEOXY-4-O-METHYL-BETA-D-ALTROPYRANOSYLOXY)METHYL]-4-FORMYL-4,4A,5,6,7,7A,8,8A-OCTAHYDRO-7-METHYL-3-(1-METHYLETHYL)-1,4-METHANO-S-INDACENE-3A(1H)-CARBOXYLIC ACID | | Authors: | Joergensen, R, Ortiz, P.A, Carr-Schmid, A, Nissen, P, Kinzy, T.G, Andersen, G.R. | | Deposit date: | 2002-10-15 | | Release date: | 2003-02-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase.
Nat. Struct. Biol., 10, 2003
|
|
4OF5
 
 | |
4OFV
 
 | |
4P39
 
 | |
1XK9
 
 | | Pseudomanas exotoxin A in complex with the PJ34 inhibitor | | Descriptor: | Exotoxin A, N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE | | Authors: | Yates, S.P, Taylor, P.J, Joergensen, R, Ferrraris, D, Zhang, J, Andersen, G.R, Merrill, A.R. | | Deposit date: | 2004-09-28 | | Release date: | 2005-05-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-function analysis of water-soluble inhibitors of the catalytic domain of exotoxin A from Pseudomonas aeruginosa
BIOCHEM.J., 385, 2005
|
|
1N0V
 
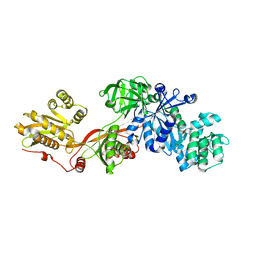 | | Crystal structure of elongation factor 2 | | Descriptor: | Elongation factor 2 | | Authors: | Joergensen, R, Ortiz, P.A, Carr-Schmid, A, Nissen, P, Kinzy, T.G, Andersen, G.R. | | Deposit date: | 2002-10-15 | | Release date: | 2002-11-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Two crystal structures demonstrate large conformational changes in the eukaryotic ribosomal translocase.
Nat.Struct.Biol., 10, 2003
|
|
8PIH
 
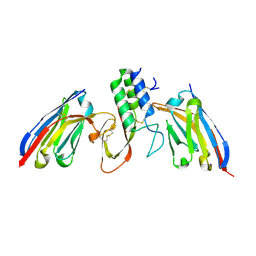 | | Structure of Api m1 in complex with two nanobodies | | Descriptor: | CHLORIDE ION, Phospholipase A2, nanobdoy AM1-1, ... | | Authors: | Aagaard, J.B, Gandini, R, Spillner, E, Miehe, M, Andersen, G.R. | | Deposit date: | 2023-06-21 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Nanobody-based IgG formats as blocking antibodies of the major honeybee venom allergen Api m 1
To be published
|
|
4P3B
 
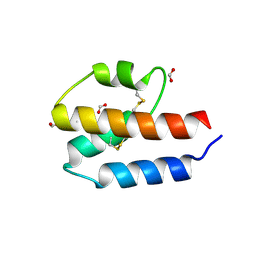 | |
4P3A
 
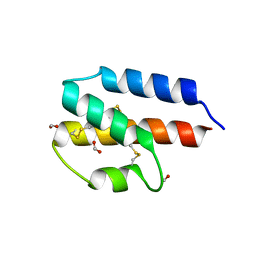 | |
4P2Y
 
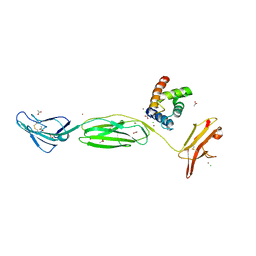 | |
1ZM3
 
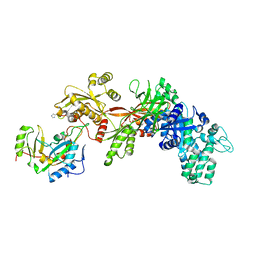 | | Structure of the apo eEF2-ETA complex | | Descriptor: | Elongation factor 2, exotoxin A | | Authors: | Joergensen, R, Merrill, A.R, Yates, S.P, Marquez, V.E, Schwan, A.L, Boesen, T, Andersen, G.R. | | Deposit date: | 2005-05-10 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry.
Nature, 436, 2005
|
|
1ZM9
 
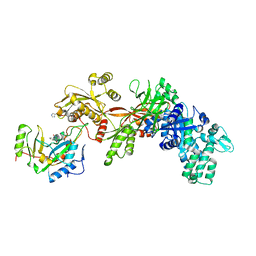 | | Structure of eEF2-ETA in complex with PJ34 | | Descriptor: | Elongation factor 2, N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, exotoxin A | | Authors: | Joergensen, R, Merrill, A.R, Yates, S.P, Marquez, V.E, Schwan, A.L, Boesen, T, Andersen, G.R. | | Deposit date: | 2005-05-10 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry.
Nature, 436, 2005
|
|
1ZM4
 
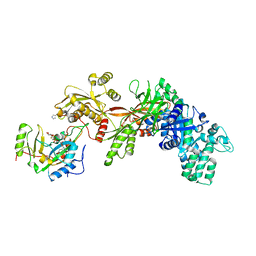 | | Structure of the eEF2-ETA-bTAD complex | | Descriptor: | BETA-METHYLENE-THIAZOLE-4-CARBOXYAMIDE-ADENINE DINUCLEOTIDE, Elongation factor 2, exotoxin A | | Authors: | Joergensen, R, Merrill, A.R, Yates, S.P, Marquez, V.E, Schwan, A.L, Boesen, T, Andersen, G.R. | | Deposit date: | 2005-05-10 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry.
Nature, 436, 2005
|
|
1ZM2
 
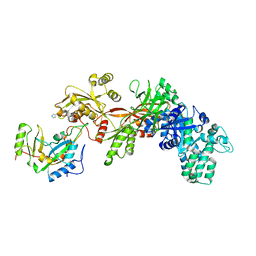 | | Structure of ADP-ribosylated eEF2 in complex with catalytic fragment of ETA | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Elongation factor 2, Exotoxin A | | Authors: | Joergensen, R, Merrill, A.R, Yates, S.P, Marquez, V.E, Schwan, A.L, Boesen, T, Andersen, G.R. | | Deposit date: | 2005-05-10 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry.
Nature, 436, 2005
|
|
7P2D
 
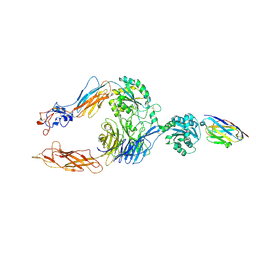 | | Structure of alphaMbeta2/Cd11bCD18 headpiece in complex with a nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jensen, R.K, Andersen, G.R. | | Deposit date: | 2021-07-05 | | Release date: | 2022-05-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the function-modulating effects of nanobody binding to the integrin receptor alpha M beta 2.
J.Biol.Chem., 298, 2022
|
|
7QIV
 
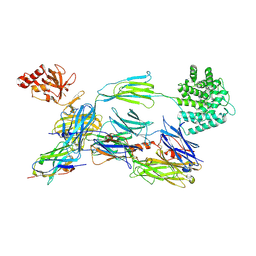 | | Structure of human C3b in complex with the EWE nanobody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3 beta chain, Complement C3b alpha' chain, ... | | Authors: | Pedersen, H, Andersen, G.R. | | Deposit date: | 2021-12-16 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Guided Engineering of a Complement Component C3-Binding Nanobody Improves Specificity and Adds Cofactor Activity.
Front Immunol, 13, 2022
|
|
7Q60
 
 | |
7Q62
 
 | |
