4YDH
 
 | | The structure of human FMNL1 N-terminal domains bound to Cdc42 | | Descriptor: | Cell division control protein 42 homolog, Formin-like protein 1, MAGNESIUM ION, ... | | Authors: | Kuhn, S, Anand, K, Geyer, M. | | Deposit date: | 2015-02-22 | | Release date: | 2015-05-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | The structure of FMNL2-Cdc42 yields insights into the mechanism of lamellipodia and filopodia formation.
Nat Commun, 6, 2015
|
|
4B2Z
 
 | | Structure of Osh6 in complex with phosphatidylserine | | Descriptor: | (2R,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, ... | | Authors: | Maeda, K, Anand, K, Chiapparino, A, Kumar, A, Poletto, M, Kaksonen, M, Gavin, A.C. | | Deposit date: | 2012-07-19 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Interactome Map Uncovers Phosphatidylserine Transport by Oxysterol-Binding Proteins
Nature, 501, 2013
|
|
7O7K
 
 | | Crystal structure of the human DYRK1A kinase domain bound to abemaciclib | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kaltheuner, I.H, Anand, K, Geyer, M. | | Deposit date: | 2021-04-13 | | Release date: | 2021-11-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Abemaciclib is a potent inhibitor of DYRK1A and HIP kinases involved in transcriptional regulation.
Nat Commun, 12, 2021
|
|
7O7I
 
 | |
7O7J
 
 | | Crystal structure of the human HIPK3 kinase domain bound to abemaciclib | | Descriptor: | Homeodomain-interacting protein kinase 3, N-{5-[(4-ethylpiperazin-1-yl)methyl]pyridin-2-yl}-5-fluoro-4-[4-fluoro-2-methyl-1-(propan-2-yl)-1H-benzimidazol-6-yl]py rimidin-2-amine | | Authors: | Kaltheuner, I.H, Anand, K, Geyer, M. | | Deposit date: | 2021-04-13 | | Release date: | 2021-11-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Abemaciclib is a potent inhibitor of DYRK1A and HIP kinases involved in transcriptional regulation.
Nat Commun, 12, 2021
|
|
1UJ1
 
 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) | | Descriptor: | 3C-like proteinase | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-07-25 | | Release date: | 2003-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1UK2
 
 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) At pH8.0 | | Descriptor: | 3C-LIKE PROTEINASE | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1UK3
 
 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) At pH7.6 | | Descriptor: | 3C-like proteinase | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1UK4
 
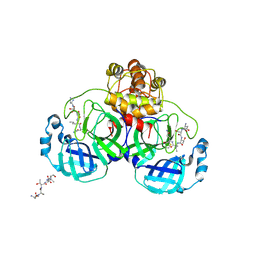 | | Crystal structure of SARS Coronavirus Main Proteinase (3CLpro) Complexed With An Inhibitor | | Descriptor: | 3C-like proteinase nsp5, 5-mer peptide of inhibitor | | Authors: | Yang, H, Yang, M, Liu, Y, Bartlam, M, Ding, Y, Lou, Z, Sun, L, Zhou, Z, Ye, S, Anand, K, Pang, H, Gao, G.F, Hilgenfeld, R, Rao, Z. | | Deposit date: | 2003-08-14 | | Release date: | 2003-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structures of severe acute respiratory syndrome virus main protease and its complex with an inhibitor
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
3ZYD
 
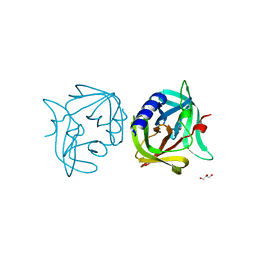 | | Crystal structure of 3C protease of coxsackievirus B3 | | Descriptor: | 3C PROTEINASE, GLYCEROL | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-22 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Peptidic Ab-Nonsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3ZYE
 
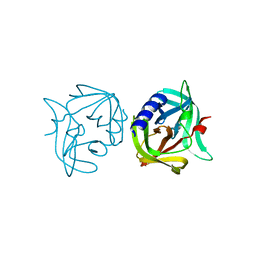 | | Crystal structure of 3C protease mutant (T68A) of coxsackievirus B3 | | Descriptor: | 3C PROTEINASE | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-22 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3ZZC
 
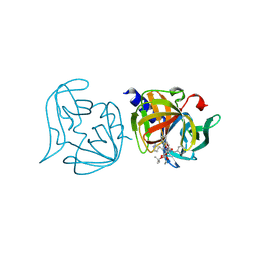 | | Crystal structure of 3C protease mutant (T68A and N126Y) of coxsackievirus B3 complexed with alpha, beta-unsaturated ethyl ester inhibitor 83 | | Descriptor: | 3C PROTEINASE, ETHYL (5S,8S,11R)-8-BENZYL-5-(2-TERT-BUTOXY-2-OXOETHYL)-3,6,9-TRIOXO-11-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}-1-PHENYL-2-OXA-4,7,10-TRIAZATETRADECAN-14-OATE | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-31 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3ZZ5
 
 | | Crystal structure of 3C protease of coxsackievirus B3 complexed with alpha, beta-unsaturated ethyl ester inhibitor 74 | | Descriptor: | 3C PROTEINASE, ETHYL (4R)-4-[(TERT-BUTOXYCARBONYL)AMINO]-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-31 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3ZZ7
 
 | | Crystal structure of 3C protease of coxsackievirus B3 complexed with alpha, beta-unsaturated ethyl ester inhibitor 81 | | Descriptor: | 3C PROTEINASE, ETHYL (4R)-4-{[N-(TERT-BUTOXYCARBONYL)-L-PHENYLALANYL]AMINO}-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-31 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3ZZ9
 
 | | Crystal structure of 3C protease of coxsackievirus B3 complexed with alpha, beta-unsaturated ethyl ester inhibitor 83 | | Descriptor: | 3C PROTEINASE, ETHYL (5S,8S,11R)-8-BENZYL-5-(2-TERT-BUTOXY-2-OXOETHYL)-3,6,9-TRIOXO-11-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}-1-PHENYL-2-OXA-4,7,10-TRIAZATETRADECAN-14-OATE | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-31 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3ZZ6
 
 | | Crystal structure of 3C protease of coxsackievirus B3 complexed with Michael receptor inhibitor 75 | | Descriptor: | ETHYL (4R)-4-({N-[(BENZYLOXY)CARBONYL]-L-PHENYLALANYL}AMINO)-5-[(3S)-2-OXOPYRROLIDIN-3-YL]PENTANOATE, POLYPROTEIN 3BCD | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-31 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Peptidic Ab-Nonsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3ZZ8
 
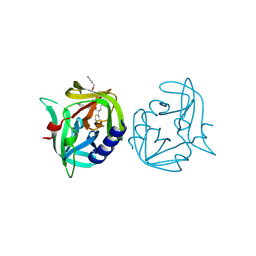 | | Crystal structure of 3C protease of coxsackievirus B3 complexed with alpha, beta-unsaturated ethyl ester inhibitor 82 | | Descriptor: | 3C PROTEINASE, ETHYL (5S,8S,11R)-8-BENZYL-5-(3-TERT-BUTOXY-3-OXOPROPYL)-3,6,9-TRIOXO-11-{[(3S)-2-OXOPYRROLIDIN-3-YL]METHYL}-1-PHENYL-2-OXA-4,7,10-TRIAZATETRADECAN-14-OATE | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-31 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3ZZA
 
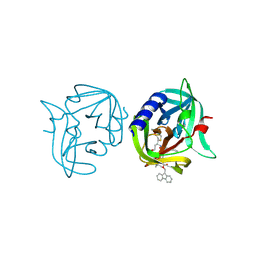 | | Crystal structure of 3C protease of coxsackievirus B3 complexed with alpha, beta-unsaturated ethyl ester inhibitor 84 | | Descriptor: | 3C PROTEINASE, O-tert-butyl-N-[(9H-fluoren-9-ylmethoxy)carbonyl]-L-threonyl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-31 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3ZZB
 
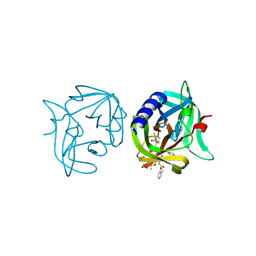 | | Crystal structure of 3C protease of coxsackievirus B3 complexed with alpha, beta-unsaturated ethyl ester inhibitor 85 | | Descriptor: | 3C PROTEINASE, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-seryl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-31 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3ZZD
 
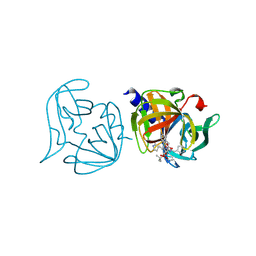 | | Crystal structure of 3C protease mutant (T68A and N126Y) of coxsackievirus B3 complexed with alpha, beta-unsaturated ethyl ester inhibitor 85 | | Descriptor: | 3C PROTEINASE, N-[(benzyloxy)carbonyl]-O-tert-butyl-L-seryl-N-{(2R)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-phenylalaninamide | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-31 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3ZZ4
 
 | | Crystal structure of 3C protease mutant (T68A and N126Y) of coxsackievirus B3 | | Descriptor: | 3C PROTEINASE | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-31 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
3ZZ3
 
 | | Crystal structure of 3C protease mutant (N126Y) of coxsackievirus B3 | | Descriptor: | 3C PROTEINASE | | Authors: | Tan, J, Anand, K, Mesters, J.R, Hilgenfeld, R. | | Deposit date: | 2011-08-31 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Peptidic Alpha, Beta-Unsaturated Ethyl Esters as Inhibitors of the 3C Protease of Coxsackie Virus B3: Crystal Structures, Antiviral Activities, and Resistance Mutations
To be Published
|
|
4WRU
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with uracil, Form II | | Descriptor: | CHLORIDE ION, GLYCEROL, URACIL, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WRW
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase, Form IV | | Descriptor: | CHLORIDE ION, GLYCEROL, Uracil-DNA glycosylase | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WRY
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-fluorouracil(B), Form I | | Descriptor: | 5-FLUOROURACIL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
