8TT7
 
 | |
1E52
 
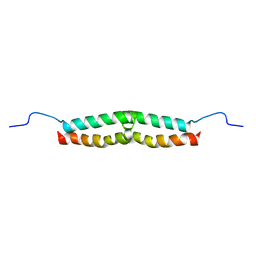 | | Solution structure of Escherichia coli UvrB C-terminal domain | | Descriptor: | EXCINUCLEASE ABC SUBUNIT | | Authors: | Alexandrovich, A.A, Kelly, G.G, Frenkiel, T.A, Moolenaar, G.F, Goosen, N.N, Sanderson, M.R, Lane, A.N. | | Deposit date: | 2000-07-14 | | Release date: | 2001-07-12 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Solution Structure, Hydrodynamics and Thermodynamics of the Uvrb C-Terminal Domain.
J.Biomol.Struct.Dyn., 19, 2001
|
|
9BV0
 
 | |
2SOB
 
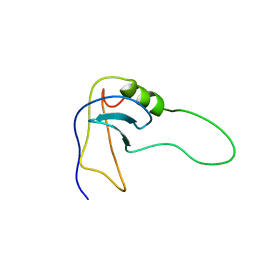 | | SN-OB, OB-FOLD SUB-DOMAIN OF STAPHYLOCOCCAL NUCLEASE, NMR, 10 STRUCTURES | | Descriptor: | STAPHYLOCOCCAL NUCLEASE | | Authors: | Alexandrescu, A.T, Gittis, A.G, Abeygunawardana, C, Shortle, D. | | Deposit date: | 1995-09-15 | | Release date: | 1995-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a stable "OB-fold" sub-domain isolated from staphylococcal nuclease.
J.Mol.Biol., 250, 1995
|
|
7OPO
 
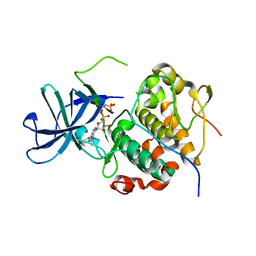 | | RSK2 N-terminal kinase domain in complex with ORF45 | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein ORF45, Ribosomal protein S6 kinase alpha-3 | | Authors: | Sok, P, Remenyi, A, Alexa, A, Poti, A. | | Deposit date: | 2021-06-01 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A non-catalytic herpesviral protein reconfigures ERK-RSK signaling by targeting kinase docking systems in the host.
Nat Commun, 13, 2022
|
|
7OPM
 
 | | Phosphorylated ERK2 in complex with ORF45 | | Descriptor: | 1-[4-(hydroxymethyl)-1H-pyrazolo[4,3-c]pyridin-6-yl]-3-[(1S)-1-phenylethyl]urea, GLYCEROL, Mitogen-activated protein kinase 1, ... | | Authors: | Sok, P, Remenyi, A, Alexa, A, Poti, A. | | Deposit date: | 2021-06-01 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A non-catalytic herpesviral protein reconfigures ERK-RSK signaling by targeting kinase docking systems in the host.
Nat Commun, 13, 2022
|
|
6WA1
 
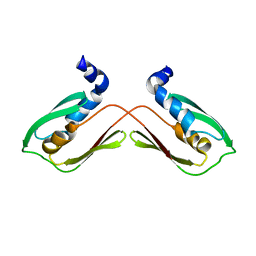 | |
1TM6
 
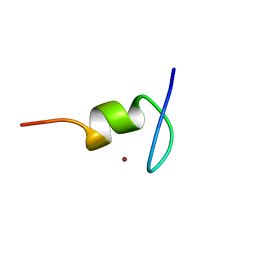 | |
8SXM
 
 | |
5IIR
 
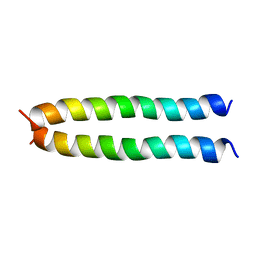 | |
6MNT
 
 | |
5IIV
 
 | | GCN4p pH 1.5 | | Descriptor: | General control protein GCN4 | | Authors: | Brady, M.R, Kaplan, A.R, Alexandrescu, A.T. | | Deposit date: | 2016-03-01 | | Release date: | 2017-03-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear Magnetic Resonance Structures of GCN4p Are Largely Conserved When Ion Pairs Are Disrupted at Acidic pH but Show a Relaxation of the Coiled Coil Superhelix.
Biochemistry, 56, 2017
|
|
5IEW
 
 | |
6E3C
 
 | |
6BEJ
 
 | | Crystal structure of manganese superoxide dismutase from Xanthomonas citri | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase | | Authors: | Goto, L.S, Alexandrino, A.V, Pereira, C.M, Mendoca, D.C, Leonardo, D.A, Pereira, H.M, Garratt, R.C, Novo-Mansur, M.T.M. | | Deposit date: | 2017-10-25 | | Release date: | 2018-10-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Structural characterization of a pathogenicity-related superoxide dismutase codified by a probably essential gene in Xanthomonas citri subsp. citri.
PLoS ONE, 14, 2019
|
|
4LH8
 
 | | Triazine hydrolase from Arthobacter aurescens modified for maximum expression in E.coli | | Descriptor: | Triazine hydrolase, ZINC ION | | Authors: | Jackson, C.J, Coppin, C.W, Alexandrov, A, Wilding, M, Liu, J.-W, Ubels, J, Paks, M, Carr, P.D, Newman, J, Russell, R.J, Field, M, Weik, M, Oakeshott, J.G, Scott, C. | | Deposit date: | 2013-07-01 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 300-Fold increase in production of the Zn2+-dependent dechlorinase TrzN in soluble form via apoenzyme stabilization.
Appl.Environ.Microbiol., 80, 2014
|
|
4L9X
 
 | | Triazine hydrolase from Arthobacter aurescens modified for maximum expression in E.coli | | Descriptor: | ACETATE ION, Triazine hydrolase | | Authors: | Jackson, C.J, Coppin, C.W, Alexandrov, A, Wilding, M, Liu, J.-W, Ubels, J, Paks, M, Carr, P.D, Newman, J, Russell, R.J, Field, M, Weik, M, Oakeshott, J.G, Scott, C. | | Deposit date: | 2013-06-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | 300-Fold increase in production of the Zn2+-dependent dechlorinase TrzN in soluble form via apoenzyme stabilization.
Appl.Environ.Microbiol., 80, 2014
|
|
6MPO
 
 | |
6D53
 
 | |
5FA7
 
 | | CTX-M-15 in complex with FPI-1523 | | Descriptor: | Beta-lactamase, [[(3~{R},6~{S})-6-(acetamidocarbamoyl)-1-methanoyl-piperidin-3-yl]amino] hydrogen sulfate | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
5FAT
 
 | | OXA-48 in complex with FPI-1602 | | Descriptor: | Beta-lactamase, CADMIUM ION, CHLORIDE ION, ... | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-12 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
5FAS
 
 | | OXA-48 in complex with FPI-1523 | | Descriptor: | Beta-lactamase, CADMIUM ION, CHLORIDE ION, ... | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
5FAO
 
 | | CTX-M-15 in complex with FPI-1465 | | Descriptor: | Beta-lactamase, [[(3~{R},6~{S})-1-methanoyl-6-[[(3~{S})-pyrrolidin-3-yl]oxycarbamoyl]piperidin-3-yl]amino] hydrogen sulfate | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
5FAQ
 
 | | OXA-48 in complex with FPI-1465 | | Descriptor: | Beta-lactamase, CADMIUM ION, CHLORIDE ION, ... | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
5FAP
 
 | | CTX-M-15 in complex with FPI-1602 | | Descriptor: | Beta-lactamase, [[(3~{R},6~{S})-6-[(azetidin-3-ylcarbonylamino)carbamoyl]-1-methanoyl-piperidin-3-yl]amino] hydrogen sulfate | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
