1FEU
 
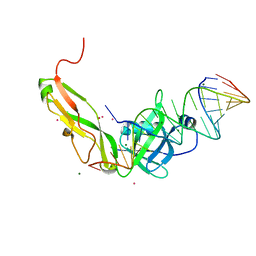 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN TL5, ONE OF THE CTC FAMILY PROTEINS, COMPLEXED WITH A FRAGMENT OF 5S RRNA. | | Descriptor: | 19 NT FRAGMENT OF 5S RRNA, 21 NT FRAGMENT OF 5S RRNA, 50S RIBOSOMAL PROTEIN L25, ... | | Authors: | Fedorov, R.V, Meshcheryakov, V.A, Gongadze, G.M, Fomenkova, N.P, Nevskaya, N.A, Selmer, M, Laurberg, M, Kristensen, O, Al-Karadaghi, S, Liljas, A, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2000-07-23 | | Release date: | 2001-06-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of ribosomal protein TL5 complexed with RNA provides new insights into the CTC family of stress proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1LD3
 
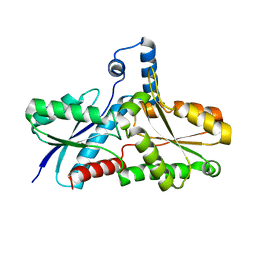 | | Crystal Structure of B. subilis ferrochelatase with Zn(2+) bound at the active site. | | Descriptor: | Ferrochelatase, ZINC ION | | Authors: | Lecerof, D, Fodje, M.N, Leon, R.A, Olsson, U, Hansson, A, Sigfridsson, E, Ryde, U, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 2002-04-08 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Metal binding to Bacillus subtilis ferrochelatase and interaction between metal sites
J.Biol.Inorg.Chem., 8, 2003
|
|
1G8P
 
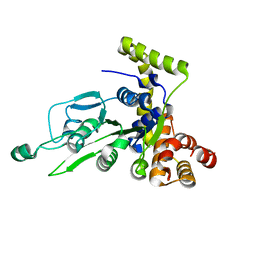 | | CRYSTAL STRUCTURE OF BCHI SUBUNIT OF MAGNESIUM CHELATASE | | Descriptor: | MAGNESIUM-CHELATASE 38 KDA SUBUNIT | | Authors: | Fodje, M.N, Hansson, A, Hansson, M, Olsen, J.G, Gough, S, Willows, R.D, Al-Karadaghi, S. | | Deposit date: | 2000-11-20 | | Release date: | 2001-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interplay between an AAA module and an integrin I domain may regulate the function of magnesium chelatase.
J.Mol.Biol., 311, 2001
|
|
2PT9
 
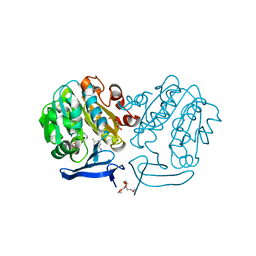 | | The structure of Plasmodium falciparum spermidine synthase in complex with decarboxylated S-adenosylmethionine and the inhibitor cis-4-methylcyclohexylamine (4MCHA) | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 5'-[(S)-(3-AMINOPROPYL)(METHYL)-LAMBDA~4~-SULFANYL]-5'-DEOXYADENOSINE, GLYCEROL, ... | | Authors: | Dufe, V.T, Qiu, W, Muller, I.B, Hui, R, Walter, R.D, Al-Karadaghi, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-08 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Plasmodium falciparum spermidine synthase in complex with the substrate decarboxylated S-adenosylmethionine and the potent inhibitors 4MCHA and AdoDATO.
J.Mol.Biol., 373, 2007
|
|
2PT6
 
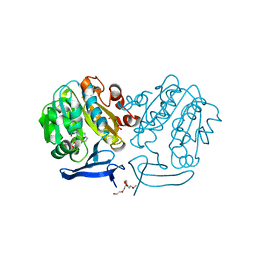 | | The structure of Plasmodium falciparum spermidine synthase in complex with decarboxylated S-adenosylmethionine | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 5'-[(S)-(3-AMINOPROPYL)(METHYL)-LAMBDA~4~-SULFANYL]-5'-DEOXYADENOSINE, GLYCEROL, ... | | Authors: | Dufe, V.T, Qiu, W, Muller, I.B, Hui, R, Walter, R.D, Al-Karadaghi, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-08 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Plasmodium falciparum spermidine synthase in complex with the substrate decarboxylated S-adenosylmethionine and the potent inhibitors 4MCHA and AdoDATO.
J.Mol.Biol., 373, 2007
|
|
2PSS
 
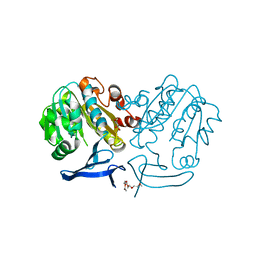 | | The structure of Plasmodium falciparum spermidine synthase in its apo-form | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, GLYCEROL, SULFATE ION, ... | | Authors: | Dufe, V.T, Qiu, W, Muller, I.B, Hui, R, Walter, R.D, Al-Karadaghi, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-07 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Plasmodium falciparum spermidine synthase in complex with the substrate decarboxylated S-adenosylmethionine and the potent inhibitors 4MCHA and AdoDATO.
J.Mol.Biol., 373, 2007
|
|
1DOZ
 
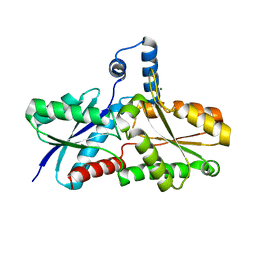 | | CRYSTAL STRUCTURE OF FERROCHELATASE | | Descriptor: | FERROCHELATASE, MAGNESIUM ION | | Authors: | Lecerof, D, Fodje, M, Hansson, A, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 1999-12-22 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mechanistic basis of porphyrin metallation by ferrochelatase.
J.Mol.Biol., 297, 2000
|
|
6SRT
 
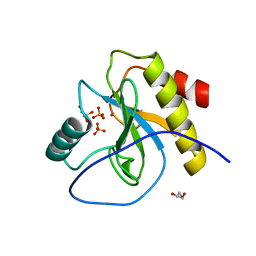 | | Endolysine N-acetylmuramoyl-L-alanine amidase LysCS from Clostridium intestinale URNW | | Descriptor: | GLYCEROL, N-acetylmuramoyl-L-alanine amidase, PHOSPHATE ION, ... | | Authors: | Hakansson, M, Al-Karadaghi, S, Plotka, M, Kaczorowska, A.-K, Kaczorowski, T. | | Deposit date: | 2019-09-06 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structure and function of endolysines LysCS, LysC from Clostridium intestinale
To Be Published
|
|
2ON3
 
 | | A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 3-aminooxy-1-aminopropane | | Descriptor: | 3-AMINOOXY-1-AMINOPROPANE, Ornithine decarboxylase | | Authors: | Dufe, V.T, Ingner, D, Heby, O, Khomutov, A.R, Persson, L, Al-Karadaghi, S. | | Deposit date: | 2007-01-23 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 1-amino-oxy-3-aminopropane.
Biochem.J., 405, 2007
|
|
2OO0
 
 | | A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 3-aminooxy-1-aminopropane | | Descriptor: | 3-AMINOOXY-1-AMINOPROPANE, ACETATE ION, Ornithine decarboxylase, ... | | Authors: | Dufe, V.T, Ingner, D, Khomutov, A.R, Heby, O, Persson, L, Al-Karadaghi, S. | | Deposit date: | 2007-01-25 | | Release date: | 2007-07-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A structural insight into the inhibition of human and Leishmania donovani ornithine decarboxylases by 1-amino-oxy-3-aminopropane.
Biochem.J., 405, 2007
|
|
2Q2O
 
 | | Crystal structure of H183C Bacillus subtilis ferrochelatase in complex with deuteroporphyrin IX 2,4-disulfonic acid dihydrochloride | | Descriptor: | Ferrochelatase, MAGNESIUM ION, PROTOPORPHYRIN IX 2,4-DISULFONIC ACID | | Authors: | Karlberg, T, Thorvaldsen, O.H, Al-Karadaghi, S. | | Deposit date: | 2007-05-29 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues
J.Mol.Biol., 378, 2008
|
|
2Q3J
 
 | | Crystal structure of the His183Ala variant of Bacillus subtilis ferrochelatase in complex with N-Methyl Mesoporphyrin | | Descriptor: | Ferrochelatase, MAGNESIUM ION, N-METHYL PROTOPORPHYRIN IX 2,4-DISULFONIC ACID | | Authors: | Karlberg, T, Thorvaldsen, O.H, Al-Karadaghi, S. | | Deposit date: | 2007-05-30 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues
J.Mol.Biol., 378, 2008
|
|
2PRL
 
 | | The structures of apo- and inhibitor bound human dihydroorotate dehydrogenase reveal conformational flexibility within the inhibitor binding site | | Descriptor: | 5-METHOXY-2-[(4-PHENOXYPHENYL)AMINO]BENZOIC ACID, ACETATE ION, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, ... | | Authors: | Walse, B, Dufe, V.T, Al-Karadaghi, S. | | Deposit date: | 2007-05-04 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structures of human dihydroorotate dehydrogenase with and without inhibitor reveal conformational flexibility in the inhibitor and substrate binding sites
Biochemistry, 47, 2008
|
|
2Q2N
 
 | | Crystal structure of Bacillus subtilis ferrochelatase in complex with deuteroporphyrin IX 2,4-disulfonic acid dihydrochloride | | Descriptor: | Ferrochelatase, MAGNESIUM ION, PROTOPORPHYRIN IX 2,4-DISULFONIC ACID | | Authors: | Karlberg, T, Thorvaldsen, O.H, Al-Karadaghi, S. | | Deposit date: | 2007-05-29 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues
J.Mol.Biol., 378, 2008
|
|
2PRM
 
 | | The structures of apo- and inhibitor bound human dihydroorotate dehydrogenase reveal conformational flexibility within the inhibitor binding site | | Descriptor: | Dihydroorotate dehydrogenase, mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Walse, B, Dufe, V.T, Al-Karadaghi, S. | | Deposit date: | 2007-05-04 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of human dihydroorotate dehydrogenase with and without inhibitor reveal conformational flexibility in the inhibitor and substrate binding sites
Biochemistry, 47, 2008
|
|
2PRH
 
 | | The structures of apo- and inhibitor bound human dihydroorotate dehydrogenase reveal conformational flexibility within the inhibitor binding site | | Descriptor: | 6-CHLORO-2-(2'-FLUOROBIPHENYL-4-YL)-3-METHYLQUINOLINE-4-CARBOXYLIC ACID, DECYLAMINE-N,N-DIMETHYL-N-OXIDE, Dihydroorotate dehydrogenase, ... | | Authors: | Walse, B, Dufe, V.T, Al-Karadaghi, S. | | Deposit date: | 2007-05-04 | | Release date: | 2008-05-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structures of human dihydroorotate dehydrogenase with and without inhibitor reveal conformational flexibility in the inhibitor and substrate binding sites
Biochemistry, 47, 2008
|
|
1BXE
 
 | | RIBOSOMAL PROTEIN L22 FROM THERMUS THERMOPHILUS | | Descriptor: | CHLORIDE ION, PROTEIN (RIBOSOMAL PROTEIN L22) | | Authors: | Unge, J, Aberg, A, Al-Karadaghi, S, Nikulin, A, Nikonov, S, Davydova, N, Nevskaya, N, Garber, M, Liljas, A. | | Deposit date: | 1998-10-02 | | Release date: | 1998-10-07 | | Last modified: | 2018-03-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of ribosomal protein L22 from Thermus thermophilus: insights into the mechanism of erythromycin resistance.
Structure, 6, 1998
|
|
