7M5A
 
 | |
7M5C
 
 | |
7M5B
 
 | |
2I9T
 
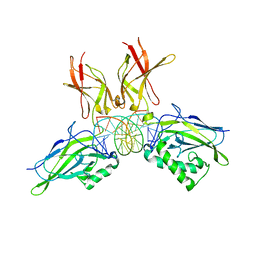 | | Structure of NF-kB p65-p50 heterodimer bound to PRDII element of B-interferon promoter | | Descriptor: | 5'-D(*AP*GP*TP*GP*GP*GP*AP*AP*AP*TP*TP*CP*CP*TP*CP*TP*G)-3', 5'-D(*CP*AP*GP*AP*GP*GP*AP*AP*TP*TP*TP*CP*CP*CP*AP*CP*T)-3', Nuclear factor NF-kappa-B p105 subunit, ... | | Authors: | Escalante, C.R, Shen, L, Thanos, D, Aggarwal, A.K. | | Deposit date: | 2006-09-06 | | Release date: | 2007-02-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of NF-kappaB p50/p65 heterodimer bound to the PRDII DNA element from the interferon-beta promoter
Structure, 10, 2002
|
|
2FOK
 
 | |
1ESG
 
 | |
6V8P
 
 | | Structure of DNA Polymerase Zeta (Apo) | | Descriptor: | DNA polymerase delta small subunit, DNA polymerase delta subunit 3, DNA polymerase zeta catalytic subunit, ... | | Authors: | Malik, R, Gomez-Llorente, Y, Ubarretxena-Belandia, I, Aggarwal, A.K. | | Deposit date: | 2019-12-11 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure and mechanism of B-family DNA polymerase zeta specialized for translesion DNA synthesis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1JIH
 
 | | Yeast DNA Polymerase ETA | | Descriptor: | DNA Polymerase ETA | | Authors: | Trincao, J, Johnson, R.E, Escalante, C.R, Prakash, S, Prakash, L, Aggarwal, A.K. | | Deposit date: | 2001-07-02 | | Release date: | 2002-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis
Mol.Cell, 8, 2001
|
|
1N72
 
 | | Structure and Ligand of a Histone Acetyltransferase Bromodomain | | Descriptor: | HISTONE ACETYLTRANSFERASE | | Authors: | Dhalluin, C, Carlson, J.E, Zeng, L, He, C, Aggarwal, A.K, Zhou, M.-M. | | Deposit date: | 2002-11-12 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and Ligand of a Histone Acetyltransferase Bromodomain
Nature, 399, 1999
|
|
1OXJ
 
 | |
2LWE
 
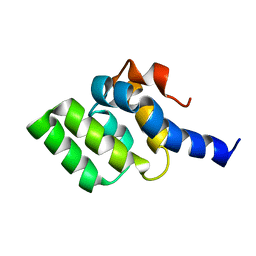 | |
2LWD
 
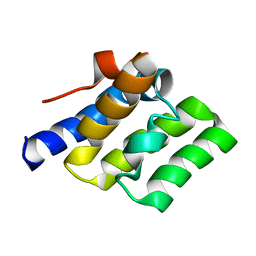 | |
6V93
 
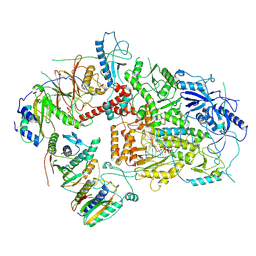 | | Structure of DNA Polymerase Zeta/DNA/dNTP Ternary Complex | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA, ... | | Authors: | Malik, R, Kopylov, M, Jain, R, Ubarrextena-Belandia, I, Aggarwal, A.K. | | Deposit date: | 2019-12-13 | | Release date: | 2020-08-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and mechanism of B-family DNA polymerase zeta specialized for translesion DNA synthesis.
Nat.Struct.Mol.Biol., 27, 2020
|
|
1MX3
 
 | | Crystal structure of CtBP dehydrogenase core holo form | | Descriptor: | ACETIC ACID, C-terminal binding protein 1, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kumar, V, Carlson, J.E, Ohgi, K.E, Edwards, T.E, Rose, D.W, Escalante, C.R, Aggarwal, A.K. | | Deposit date: | 2002-10-01 | | Release date: | 2002-12-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Transcription Corepressor CtBP Is an NAD+-Regulated Dehydrogenase
Mol.Cell, 10, 2002
|
|
4ZCF
 
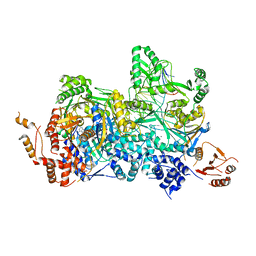 | | Structural basis of asymmetric DNA methylation and ATP-triggered long-range diffusion by EcoP15I | | Descriptor: | ADENOSINE MONOPHOSPHATE, CALCIUM ION, DNA 20-mer AATCATAGTCTACTGCTGTA, ... | | Authors: | Gupta, Y.K, Chan, S.H, Xu, S.Y, Aggarwal, A.K. | | Deposit date: | 2015-04-15 | | Release date: | 2015-07-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of asymmetric DNA methylation and ATP-triggered long-range diffusion by EcoP15I.
Nat Commun, 6, 2015
|
|
1JGG
 
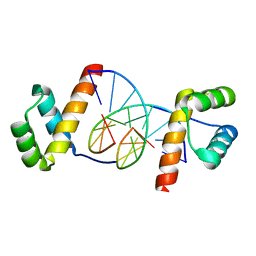 | | Even-skipped Homeodomain Complexed to AT-rich DNA | | Descriptor: | 5'-D(P*AP*AP*TP*TP*CP*AP*AP*TP*TP*A)-3', 5'-D(P*TP*AP*AP*TP*TP*GP*AP*AP*TP*T)-3', Segmentation Protein Even-Skipped | | Authors: | Hirsch, J.A, Aggarwal, A.K. | | Deposit date: | 2001-06-25 | | Release date: | 2001-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the even-skipped homeodomain complexed to AT-rich DNA: new perspectives on homeodomain specificity.
EMBO J., 14, 1995
|
|
1Q7F
 
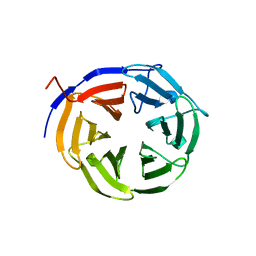 | |
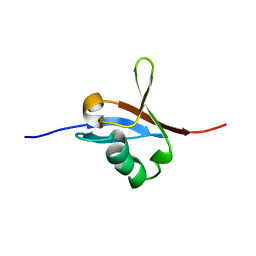
5J17
 
 | |
5KQR
 
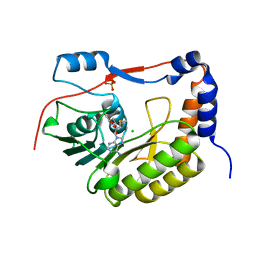 | | Structure of NS5 methyltransferase from Zika virus bound to S-adenosylmethionine | | Descriptor: | CHLORIDE ION, Methyltransferase, PHOSPHATE ION, ... | | Authors: | Jain, R, Coloma, J, Rajashankar, K.R, Aggarwal, A.K. | | Deposit date: | 2016-07-06 | | Release date: | 2016-09-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.331 Å) | | Cite: | Structures of NS5 Methyltransferase from Zika Virus.
Cell Rep, 16, 2016
|
|
1ES8
 
 | | Crystal structure of free BglII | | Descriptor: | ACETIC ACID, RESTRICTION ENDONUCLEASE BGLII | | Authors: | Lukacs, C.M, Aggarwal, A.K. | | Deposit date: | 2000-04-07 | | Release date: | 2001-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of free BglII reveals an unprecedented scissor-like motion for opening an endonuclease.
Nat.Struct.Biol., 8, 2001
|
|
5J2R
 
 | | Solution structure of Ras Binding Domain (RBD) of B-Raf | | Descriptor: | N-[2-methoxy-5-({[(E)-2-(2,4,6-trimethoxyphenyl)ethenyl]sulfonyl}methyl)phenyl]glycine, Serine/threonine-protein kinase B-raf | | Authors: | Dutta, K, Vasquez-Del Carpio, R, Aggarwal, A.K, Reddy, E.P. | | Deposit date: | 2016-03-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule RAS-Mimetic Disrupts RAS Association with Effector Proteins to Block Signaling.
Cell, 165, 2016
|
|
6W32
 
 | | Crystal structure of Sfh5 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Phosphatidylinositol transfer protein SFH5 | | Authors: | Gulten, G, Khan, D, Aggarwal, A, Krieger, I, Sacchettini, J.C, Bankaitis, V.A. | | Deposit date: | 2020-03-08 | | Release date: | 2020-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Sec14-like phosphatidylinositol transfer protein paralog defines a novel class of heme-binding proteins.
Elife, 9, 2020
|
|
1D2I
 
 | | CRYSTAL STRUCTURE OF RESTRICTION ENDONUCLEASE BGLII COMPLEXED WITH DNA 16-MER | | Descriptor: | DNA (5'-D(*TP*AP*TP*TP*AP*TP*AP*GP*AP*TP*CP*TP*AP*TP*AP*A)-3'), MAGNESIUM ION, PROTEIN (RESTRICTION ENDONUCLEASE BGLII) | | Authors: | Lukacs, C.M, Kucera, R, Schildkraut, I, Aggarwal, A.K. | | Deposit date: | 1999-09-23 | | Release date: | 2000-02-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Understanding the immutability of restriction enzymes: crystal structure of BglII and its DNA substrate at 1.5 A resolution.
Nat.Struct.Biol., 7, 2000
|
|
5L2X
 
 | | Crystal structure of human PrimPol ternary complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(P*GP*GP*TP*AP*GP*CP*(DDG))-3'), ... | | Authors: | Rechkoblit, O, Gupta, Y.K, Malik, R, Rajashankar, K.R, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2016-08-02 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and mechanism of human PrimPol, a DNA polymerase with primase activity.
Sci Adv, 2, 2016
|
|
2FLN
 
 | | binary complex of catalytic core of human DNA polymerase iota with DNA (template A) | | Descriptor: | DNA polymerase iota, DNA primer strand, DNA template strand | | Authors: | Nair, D.T, Johnson, R.E, Prakash, L, Prakash, S, Aggarwal, A.K. | | Deposit date: | 2006-01-06 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An incoming nucleotide imposes an anti to syn conformational change on the templating purine in the human DNA polymerase-iota active site.
Structure, 14, 2006
|
|
