8C7J
 
 | |
8C7V
 
 | |
5FUC
 
 | | Biophysical and cellular characterisation of a junctional epitope antibody that locks IL-6 and gp80 together in a stable complex: implications for new therapeutic strategies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, INTERLEUKIN-6, INTERLEUKIN-6 RECEPTOR SUBUNIT ALPHA, ... | | Authors: | Adams, R, Griffin, R, Doyle, C, Ettorre, A. | | Deposit date: | 2016-01-25 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of a junctional epitope antibody that stabilizes IL-6 and gp80 protein:protein interaction and modulates its downstream signaling.
Sci Rep, 7, 2017
|
|
5FUO
 
 | |
5FUZ
 
 | |
8RUU
 
 | | Fabs derived from bimekizumab in complex with IL-17F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoblobulin heavy chain, Immunoblobulin light chain, ... | | Authors: | Adams, R, Lawson, A.D.G. | | Deposit date: | 2024-01-31 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Crystal Structure of Bimekizumab Fab Fragment in Complex with IL-17F Provides Molecular Basis for Dual IL-17A and IL-17F Inhibition.
J Invest Dermatol., 2024
|
|
5NLB
 
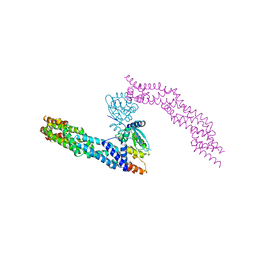 | | Crystal structure of human CUL3 N-terminal domain bound to KEAP1 BTB and 3-box | | Descriptor: | Cullin-3, Kelch-like ECH-associated protein 1 | | Authors: | Adamson, R, Krojer, T, Pinkas, D.M, Bartual, S.G, Burgess-Brown, N.A, Borkowska, O, Chalk, R, Newman, J.A, Kopec, J, Dixon-Clarke, S.E, Mathea, S, Sethi, R, Velupillai, S, Mackinnon, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2017-04-04 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Structural and biochemical characterization establishes a detailed understanding of KEAP1-CUL3 complex assembly.
Free Radic Biol Med, 204, 2023
|
|
6Z36
 
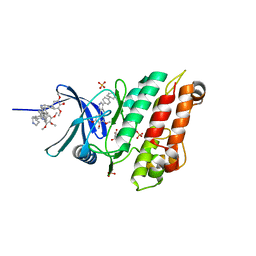 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2118 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-(4-piperidin-4-ylphenyl)-5-(3,4,5-trimethoxyphenyl)pyridine, AMMONIUM ION, ... | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-05-19 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2118
To Be Published
|
|
6SZM
 
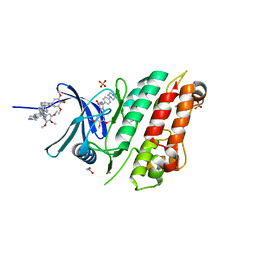 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2009 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-[4-methyl-5-(3,4,5-trimethoxyphenyl)pyridin-3-yl]phenyl]piperazine, AMMONIUM ION, ... | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-10-02 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Leveraging an Open Science Drug Discovery Model to Develop CNS-Penetrant ALK2 Inhibitors for the Treatment of Diffuse Intrinsic Pontine Glioma.
J.Med.Chem., 63, 2020
|
|
6T6D
 
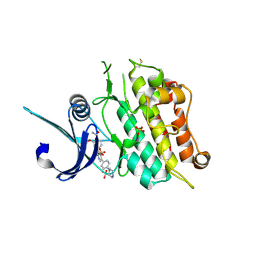 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2149 | | Descriptor: | 2-methoxy-4-[4-methyl-5-(4-piperazin-1-ylphenyl)pyridin-3-yl]benzamide, Activin receptor type I, SULFATE ION | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-10-18 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Targeting ALK2: An Open Science Approach to Developing Therapeutics for the Treatment of Diffuse Intrinsic Pontine Glioma.
J.Med.Chem., 63, 2020
|
|
6SRH
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2117 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type-1, ... | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-09-05 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2117
To Be Published
|
|
7A21
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2158 | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(pyrrolidin-1-ylmethyl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type I, ... | | Authors: | Adamson, R.J, Williams, E.P, Smil, D, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-08-14 | | Release date: | 2020-09-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2153
To Be Published
|
|
6T8N
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K3007 | | Descriptor: | 1,2-ETHANEDIOL, Activin receptor type I, DIMETHYL SULFOXIDE, ... | | Authors: | Adamson, R.J, Williams, E.P, Bonomo, S, Rankin, S, Bacos, D, Rae, A, Cramp, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-10-24 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K3007
To Be Published
|
|
5F72
 
 | | De novo design and crystallographic validation of antibodies targeting a pre-selected epitope | | Descriptor: | Kelch-like ECH-associated protein 1, Single chain Fv from a Fab | | Authors: | Liu, X, Taylor, R.D, Griffin, L, Coker, S, Adams, R, Ceska, T, Shi, J, Lawson, A.D.G, Baker, T. | | Deposit date: | 2015-12-07 | | Release date: | 2016-12-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | De novo design and crystallographic validation of antibodies targeting a pre-selected epitope
To Be Published
|
|
7EAD
 
 | | Crystal structure of beta-sheet cytochrome c prime from Thermus thermophilus. | | Descriptor: | Cytochrome_P460 domain-containing protein, HEME C | | Authors: | Yoshimi, T, Fujii, S, Oki, H, Igawa, T, Adams, R.H, Ueda, K, Kawahara, K, Ohkubo, T, Hough, A.M, Sambongi, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of thermally stable homodimeric cytochrome c'-beta from Thermus thermophilus.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
4CNI
 
 | | Crystal structure of the Fab portion of Olokizumab in complex with IL- 6 | | Descriptor: | INTERLEUKIN-6, OLOKIZUMAB HEAVY CHAIN, FAB PORTION, ... | | Authors: | Shaw, S, Bourne, T, Meier, C, Carrington, B, Gelinas, R, Henry, A, Popplewell, A, Adams, R, Baker, T, Rapecki, S, Marshall, D, Neale, H, Lawson, A. | | Deposit date: | 2014-01-22 | | Release date: | 2014-04-30 | | Last modified: | 2019-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Characterization of Olokizumab: A Humanized Antibody Targeting Interleukin-6 and Neutralizing Gp130-Signaling.
Mabs, 6, 2014
|
|
5L6W
 
 | | Structure Of the LIMK1-ATPgammaS-CFL1 Complex | | Descriptor: | Cofilin-1, LIM domain kinase 1, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Salah, E, Mathea, S, Oerum, S, Newman, J.A, Tallant, C, Adamson, R, Canning, P, Beltrami, A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Bullock, A.N. | | Deposit date: | 2016-05-31 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structure Of the LIMK1-ATPgammaS-CFL1 Complex
To Be Published
|
|
5G3Q
 
 | | Crystal structure of a hypothetical domain in WNK1 | | Descriptor: | WNK1 | | Authors: | Pinkas, D.M, Bufton, J.C, Sanvitale, C.E, Bartual, S.G, Adamson, R.J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2016-04-29 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.613 Å) | | Cite: | Crystal Structure of a Hypothetical Domain in Wnk1
To be Published
|
|
3L0H
 
 | | Crystal Structure Analysis of W21A mutant of human GSTA1-1 in complex with S-hexylglutathione | | Descriptor: | Glutathione S-transferase A1, S-HEXYLGLUTATHIONE | | Authors: | Fanucchi, S, Achilonu, I.A, Adamson, R.J, Fernandes, M.A, Burke, J.P, Dirr, H.W. | | Deposit date: | 2009-12-10 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Stability of the domain interface contributes towards the catalytic function at the H-site of class alpha glutathione transferase A1-1.
Biochim.Biophys.Acta, 1804, 2010
|
|
3O3T
 
 | | Crystal Structure Analysis of M32A mutant of human CLIC1 | | Descriptor: | Chloride intracellular channel protein 1 | | Authors: | Fanucchi, S, Achilonu, I.A, Adamson, R.J, Fernandes, M.A, Stoychev, S, Dirr, H.W. | | Deposit date: | 2010-07-26 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure Analysis of M32A mutant of human CLIC1
To be Published
|
|
5S7N
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010920a | | Descriptor: | 1,2-ETHANEDIOL, 1H-imidazol-2-amine, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S7W
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with HM000007h | | Descriptor: | 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, Activin receptor type-1, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S7Y
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010933a | | Descriptor: | (2S,4R)-1,3-thiazolidine-2,4-diamine, 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S84
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with FM010949a | | Descriptor: | 1,2-ETHANEDIOL, 1-[(4R)-1,3-oxazolidin-4-yl]methanamine, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | XChem group deposition
To Be Published
|
|
5S8A
 
 | | XChem group deposition -- Crystal Structure of human ACVR1 in complex with NU074484b | | Descriptor: | (4S)-imidazolidine-4-carbonitrile, 1,2-ETHANEDIOL, 4-methyl-3-[4-(1-methylpiperidin-4-yl)phenyl]-5-(3,4,5-trimethoxyphenyl)pyridine, ... | | Authors: | Williams, E.P, Adamson, R.J, Smil, D, Krojer, T, Burgess-Brown, N, von Delft, F, Bountra, C, Bullock, A.N. | | Deposit date: | 2020-12-11 | | Release date: | 2021-06-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | XChem group deposition
To Be Published
|
|
